| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,010,858 – 3,010,978 |
| Length | 120 |
| Max. P | 0.550215 |

| Location | 3,010,858 – 3,010,978 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.14 |
| Shannon entropy | 0.56338 |
| G+C content | 0.50385 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -17.80 |
| Energy contribution | -16.52 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.96 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
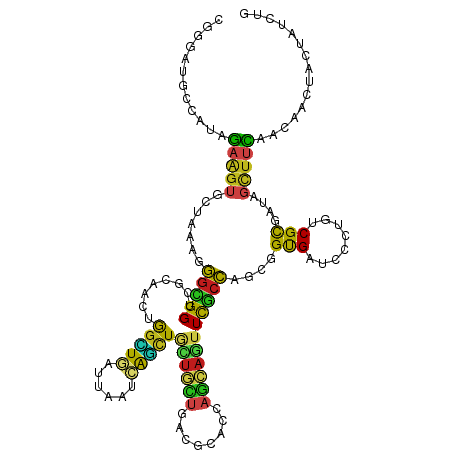
>dm3.chr3R 3010858 120 - 27905053 CGGGAUGCCAUAGAAGUUUUAAAGGGCGGUCGCAACUGGUUAAUUAAUCAGCUGCUGCUAACGCACCAGCAGUUCGCCAGCAGUGAUCCCAGUCGCGAUAGCUUCAACAACUACUAUUUG .((....))((((.((((.....((((.(((((.(((((...........((((.(((....))).))))...((((.....))))..))))).))))).))))....)))).))))... ( -38.40, z-score = -2.10, R) >droSim1.chr3R 3053146 120 - 27517382 CGGGAUGCCAUAGAAGUUUUAAAGGGCGGUCGCAACUGGUUAAUUAACCAGCUGCUGCUAACGCACCAGCAGUUCGCCAGCAGUGAUCCCAGUCGCGAUAGCUUCAACAACUACUAUUUG .((....))((((.((((.....((((.(((((.(((((...........((((.(((....))).))))...((((.....))))..))))).))))).))))....)))).))))... ( -38.40, z-score = -1.98, R) >droSec1.super_6 3103040 120 - 4358794 CGGGAUGCCAUAGAAGUUUUAAAGGGCGGUCGCAACUGGUUAAUUAACCAGCUGCUGCUAACGCACCAGCAGUUCGCCAGCAGUGAUCCCAGUCGCGAUAGCUUCAACAACUACUAUUUG .((....))((((.((((.....((((.(((((.(((((...........((((.(((....))).))))...((((.....))))..))))).))))).))))....)))).))))... ( -38.40, z-score = -1.98, R) >droYak2.chr3R 21502793 120 + 28832112 CGAGAUGCCAUAGAAGUUUUAAAGGGCGGGCGCAACUGGUUGAUUAACCAGUUGCUGCUGACGCACCAGCAGUUCGCCAGUGGUGAUCCCAGUCGCGAUAGCUUCAACAGUUACUAUUUG ............((((((......(((((((((((((((((....))))))))))(((((......)))))))))))).((.(((((....))))).))))))))............... ( -44.90, z-score = -3.18, R) >droEre2.scaffold_4770 3273365 120 - 17746568 CGGGAUGCCAUAGAAGUUUUAAAGGGCGGGCGAAACUGGUUGAUUAACCAGCUGCUGCUGACACACCAGCAGUUUGCCAGCAGUGAACCUAGUCGCGAUAGCUUCAACAGCUACUAUUUG .((...((....((((((.......((.(((((..((((((....))))))..(((((((......)))))))))))).)).((((......))))...))))))....))..))..... ( -36.30, z-score = -0.67, R) >droAna3.scaffold_13340 2784466 120 - 23697760 CGAGAUGCUAUCGAGGUGCUGAAGGGUGGCCGCAACUGGCUGAUCAAUCAACUACUUCUCACACACCAGCAGUUCGCCAGUGGCGAUCCCUGUCGCGACAGCUUUAACAAGUACUACCUG ......(((.(((.((((.(((..((((((((....))))(((....)))..))))..)))..)))).((((.(((((...)))))...))))..))).))).................. ( -32.00, z-score = 0.68, R) >dp4.chr2 26422091 120 - 30794189 CGGGAUGCCAUCGAGGUGCUAAAAGGCGGUCGGAACUGGCUGAUUAAUCAGCUUUUGUUGACGCACCAGCAGUUUGCCAGCGGUGAGCCGUGCCGCGAGAGCUAUAACACCUACUACCUG ((((.........(((((......((((((..((((((.(((.....(((((....))))).....)))))))))))).((((....)))))))((....)).....)))))....)))) ( -39.62, z-score = -0.23, R) >droPer1.super_6 1750786 120 - 6141320 CGGGAUGCCAUCGAGGUGCUAAAAGGCGGUCGGAACUGGCUGAUUAAUCAGCUUUUGUUGACGCACCAGCAGUUUGCCAGCGGUGAGCCGUGCCGCGAGAGCUACAACACCUACUACCUG ((((.........(((((......((((((..((((((.(((.....(((((....))))).....)))))))))))).((((....)))))))((....)).....)))))....)))) ( -39.62, z-score = -0.14, R) >droWil1.scaffold_181130 16617170 120 - 16660200 CGUGAUGCCAUUGAGGUGCUCAGGGGAGGAAGAAAUUGGUUGAUUAAUCAAUUACUACUGACCCAUCAACAAUUUGCCACUAGUGAACCGAGCCGCGAUACCUUUAAUAGCCUUUAUAUA .((((.((.((((((((((((.(((..((.((.((((((((....)))))))).)).))..)))........((..(.....)..))..))))......))).))))).))..))))... ( -28.20, z-score = -0.85, R) >droMoj3.scaffold_6540 20871742 120 + 34148556 AGGGAUGCCAUUGAAGUACUGAGGGGUGGUCGCAACUGGCUGAUCAAUCAGCUGUUGCUGACCCACCUGCAGUUCGCCAGCGGCGAGCCAUGUCGCGAAAGCUUCAACAACUAUUAUCUG .((..((((.(((..(.((((..(((((((((((((.((((((....))))))))))).)).)))))).)))).)..))).))))..)).((..((....))..)).............. ( -46.80, z-score = -2.33, R) >droGri2.scaffold_14906 3303907 120 - 14172833 CGGGAUGCCAUCGAGGUGCUGAAGGGCGGUCGCAAUUGGCUGAUCAAUCAGCUGCUGUUGACCCACCAGCAAUUUGCCAGUGGCGAACCGUGUCGCGAUAGUUUCAACAGCCAUUAUCUG (((..((((((((((.(((((..((((((..(((....(((((....))))))))..))).)))..))))).))))...))))))..)))......((((((..(....)..)))))).. ( -40.10, z-score = -0.22, R) >droVir3.scaffold_13047 16861165 120 - 19223366 AGGGAUGCCAUCGAAGUACUGAAGGGUGGACGAAACUGGCUGAUCAGUCAGCUGCUGCUGACCCACCAGCAGUUCGCCAGUGGCGAGCCAUGUCGCGAUAGUUUCAACAACUAUUAUCUG ..((((((((((............)))))..((((((((((((....))))))(((((((......)))))))((((..((((....))))...)))).)))))).........))))). ( -41.10, z-score = -1.18, R) >triCas2.ChLG2 3753701 120 - 12900155 ACCGAUUUGAGGAAACUACUCGAUUUGGGCGAAAAAUAUAUAAUCAAGUGUUUGAUUGAAAUGUACCACAUUUUUUUGACUGGCGAUUGUUGCCGUUACAUUCUUAACAAUUUGUUCAUU .((((.(((((.......))))).))))(((((.......(((((((....))))))).((((((.(.((......))...(((((...)))))).))))))........)))))..... ( -23.70, z-score = -0.67, R) >consensus CGGGAUGCCAUAGAAGUGCUAAAGGGCGGUCGCAACUGGCUGAUUAAUCAGCUGCUGCUGACGCACCAGCAGUUCGCCAGCGGUGAUCCCUGUCGCGAUAGCUUCAACAACUACUAUCUG ............(((((.......(((((........(((((......)))))((((((........)))))))))))....(((........)))....)))))............... (-17.80 = -16.52 + -1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:32 2011