| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,975,666 – 2,975,760 |
| Length | 94 |
| Max. P | 0.921447 |

| Location | 2,975,666 – 2,975,760 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 59.65 |
| Shannon entropy | 0.72032 |
| G+C content | 0.33185 |
| Mean single sequence MFE | -16.21 |
| Consensus MFE | -7.02 |
| Energy contribution | -7.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
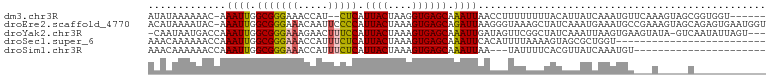
>dm3.chr3R 2975666 94 + 27905053 AUAUAAAAAAC-AAAUUGGCGGGAAACCAU--CUCAUUACUAAGGUGAGCAAAUUAACCUUUUUUUUACAUUAUCAAAUGUUCAAAGUAGCGGUGGU------ ...........-........((....))..--.((((..(((...((((((...........................))))))...)))..)))).------ ( -15.83, z-score = -0.67, R) >droEre2.scaffold_4770 3237959 102 + 17746568 ACAUAAAAUAC-AAAUUGGCGGGAAACAAUUCCCCAUUACUAAAGUGAGCAGAUUAAGGGUAAAGCUAUCAAAUGAAAUGCCGAAAGUAGCAGAGUGAAUGGU .(((....(((-...((((((((((....)))))..(((((....(((.....)))..)))))................)))))..))).....)))...... ( -17.30, z-score = -0.12, R) >droYak2.chr3R 21455551 98 - 28832112 -CAAUAAUGACCAAAUUGGCGGGAAAGAACUUUCCAUUACUAAAGUGAGCAAAUUGAUAGUUCGGCUAUCAAAUUAAGUGAAGUAUA-GUCAAUAUUAGU--- -.((((.((((.......((.(((((....))))).((((....))))))...(((((((.....)))))))...............-)))).))))...--- ( -20.70, z-score = -2.12, R) >droSec1.super_6 3068101 78 + 4358794 AAACAAAAAACCAAAUUGGCGGGAAACCAUUUCUCAUUACUAAAGUGAGCAAAUUCACAUUUUAAAAGUAGCGCUGGU------------------------- ..............((..((((....))((((((((((.....)))))).))))..................))..))------------------------- ( -15.50, z-score = -1.85, R) >droSim1.chr3R 3015709 78 + 27517382 AAACAAAAAACCAAAUUGGCGGGAAACCAUUUCUCAUUACUAAAGUGAGCAAAUUAA---UAUUUUCACGUUAUCAAAUGU---------------------- ................((((((....))((((((((((.....)))))).))))...---.........))))........---------------------- ( -11.70, z-score = -1.50, R) >consensus AAACAAAAAACCAAAUUGGCGGGAAACCAUUUCUCAUUACUAAAGUGAGCAAAUUAACAGUUUAGCUAUAAAAUCAAAUGU___A_____C____________ .............((((.(((((((.....))))).((((....)))))).))))................................................ ( -7.02 = -7.06 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:26 2011