
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,956,655 – 2,956,744 |
| Length | 89 |
| Max. P | 0.990849 |

| Location | 2,956,655 – 2,956,744 |
|---|---|
| Length | 89 |
| Sequences | 13 |
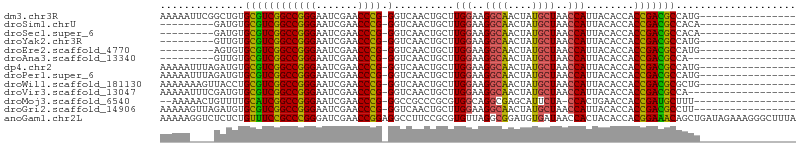
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.40259 |
| G+C content | 0.55142 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
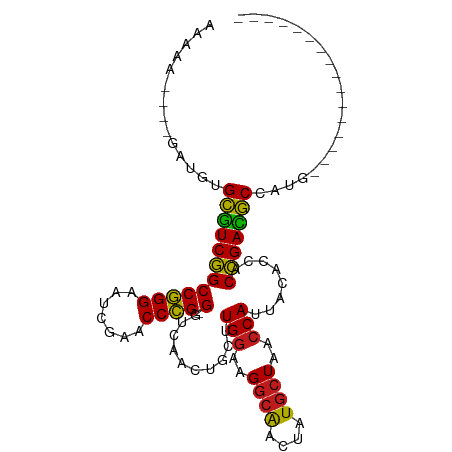
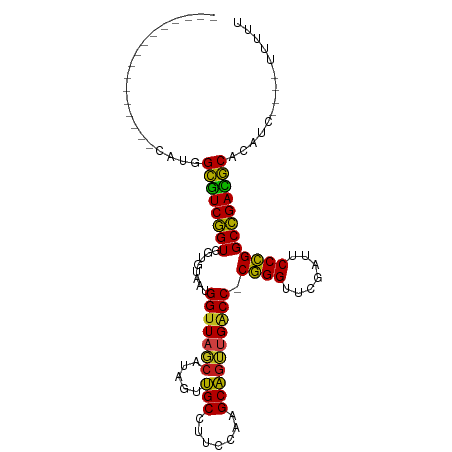
>dm3.chr3R 2956655 89 + 27905053 AAAAAUUCGGCUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCAUG---------------- ...........((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))..---------------- ( -29.10, z-score = -1.68, R) >droSim1.chrU 9968918 80 + 15797150 ---------GAUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCACA---------------- ---------..(((((((((((((((.......))))-)..........(((..((((....))))..)))........)))))).))))---------------- ( -29.70, z-score = -2.63, R) >droSec1.super_6 3046616 80 + 4358794 ---------GAUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCACA---------------- ---------..(((((((((((((((.......))))-)..........(((..((((....))))..)))........)))))).))))---------------- ( -29.70, z-score = -2.63, R) >droYak2.chr3R 21437683 80 - 28832112 ---------GUUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCAUG---------------- ---------..((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))..---------------- ( -29.20, z-score = -2.22, R) >droEre2.scaffold_4770 3217604 80 + 17746568 ---------AGUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCAUG---------------- ---------.(((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))).---------------- ( -30.80, z-score = -2.57, R) >droAna3.scaffold_13340 2735089 78 + 23697760 ---------GUUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCA------------------ ---------..((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))------------------ ( -28.60, z-score = -1.97, R) >dp4.chr2 26370607 89 + 30794189 AAAAAUUUAGAUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCAUG---------------- ..........(((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))).---------------- ( -30.60, z-score = -2.95, R) >droPer1.super_6 1700069 89 + 6141320 AAAAAUUUAGAUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCAUG---------------- ..........(((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))).---------------- ( -30.60, z-score = -2.95, R) >droWil1.scaffold_181130 16556244 89 + 16660200 AAAAAAAGUUACCUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCGCUG---------------- ..............((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))....---------------- ( -29.00, z-score = -2.23, R) >droVir3.scaffold_13047 16808559 87 + 19223366 AAAAAUUUCGAUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCA------------------ ...........((.((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))))------------------ ( -28.70, z-score = -2.08, R) >droMoj3.scaffold_6540 11342119 85 - 34148556 --AAAAACUGUUUUGCAUCGGCCGGGAAUCGAACCCG-GGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUU----------------- --............((((((((((((.......))))-)((((((.((...)).)))).))......-...........)))))))...----------------- ( -32.20, z-score = -1.89, R) >droGri2.scaffold_14906 3240231 88 + 14172833 AAAAAGUUAGAUGUGCGUCGGCCGGGAAUCGAACCCG-GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCUU----------------- ..............((((((((((((.......))))-)..........(((..((((....))))..)))........)))))))...----------------- ( -28.10, z-score = -1.81, R) >anoGam1.chr2L 33883185 106 - 48795086 AAAAAGGUCUCUCUGUUUCCGCCCGGGAUCGAACCGGAGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACAGCUGAUAGAAAGGGCUUUA ...((((((((((((((((((.((((.......)))).((...(((((.......))))).(((.....)))....)).))))))))).....)))...)))))). ( -34.30, z-score = -0.89, R) >consensus AAAAA____GAUGUGCGUCGGCCGGGAAUCGAACCCG_GGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCAUG________________ ..............(((((((.((((.......)))).(((........(((..((((....))))..)))....))).))))))).................... (-23.63 = -23.18 + -0.44)
| Location | 2,956,655 – 2,956,744 |
|---|---|
| Length | 89 |
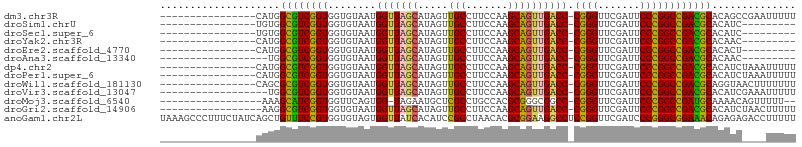
| Sequences | 13 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.40259 |
| G+C content | 0.55142 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -28.97 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2956655 89 - 27905053 ----------------CAUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAGCCGAAUUUUU ----------------..((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))........... ( -30.90, z-score = -1.39, R) >droSim1.chrU 9968918 80 - 15797150 ----------------UGUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAUC--------- ----------------.(((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))).--------- ( -31.50, z-score = -2.11, R) >droSec1.super_6 3046616 80 - 4358794 ----------------UGUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAUC--------- ----------------.(((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))).--------- ( -31.50, z-score = -2.11, R) >droYak2.chr3R 21437683 80 + 28832112 ----------------CAUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAAC--------- ----------------..((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))..--------- ( -30.90, z-score = -2.06, R) >droEre2.scaffold_4770 3217604 80 - 17746568 ----------------CAUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACACU--------- ----------------..((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))..--------- ( -30.60, z-score = -1.77, R) >droAna3.scaffold_13340 2735089 78 - 23697760 ------------------UGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAAC--------- ------------------((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))..--------- ( -30.60, z-score = -2.02, R) >dp4.chr2 26370607 89 - 30794189 ----------------CAUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAUCUAAAUUUUU ----------------.(((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))).......... ( -31.30, z-score = -2.20, R) >droPer1.super_6 1700069 89 - 6141320 ----------------CAUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAUCUAAAUUUUU ----------------.(((((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).))).......... ( -31.30, z-score = -2.20, R) >droWil1.scaffold_181130 16556244 89 - 16660200 ----------------CAGCGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCAGGUAACUUUUUUU ----------------....((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).............. ( -30.70, z-score = -1.61, R) >droVir3.scaffold_13047 16808559 87 - 19223366 ------------------UGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAUCGAAAUUUUU ------------------..((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).............. ( -30.40, z-score = -1.68, R) >droMoj3.scaffold_6540 11342119 85 + 34148556 -----------------AAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCC-CGGGUUCGAUUCCCGGCCGAUGCAAAACAGUUUUU-- -----------------...(((((((..((((.....-..))))((.(((((((...)))))))))(-((((.......))))))))))))............-- ( -35.90, z-score = -1.94, R) >droGri2.scaffold_14906 3240231 88 - 14172833 -----------------AAGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC-CGGGUUCGAUUCCCGGCCGACGCACAUCUAACUUUUU -----------------...((((((((........(((((((...(((........))).)))))))-((((.......)))))))))))).............. ( -30.40, z-score = -1.95, R) >anoGam1.chr2L 33883185 106 + 48795086 UAAAGCCCUUUCUAUCAGCUGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCUCCGGUUCGAUCCCGGGCGGAAACAGAGAGACCUUUUU ..........(((.....(((((((((((((.(.(((.....))).(((((.......)))))).))).((((.......))))))))))))))..)))....... ( -35.00, z-score = -1.49, R) >consensus ________________CAUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACC_CGGGUUCGAUUCCCGGCCGACGCACAUC____UUUUU ....................((((((((........(((((((.....(((.......)))))))))).((((.......)))))))))))).............. (-28.97 = -28.20 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:24 2011