
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,850,218 – 2,850,322 |
| Length | 104 |
| Max. P | 0.778464 |

| Location | 2,850,218 – 2,850,322 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.11 |
| Shannon entropy | 0.20854 |
| G+C content | 0.46845 |
| Mean single sequence MFE | -29.59 |
| Consensus MFE | -23.93 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
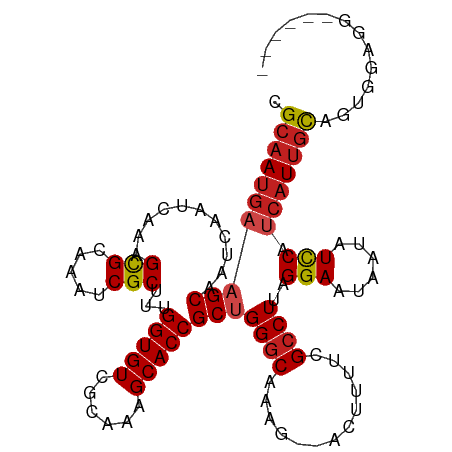
>dm3.chr3R 2850218 104 + 27905053 CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGG------ .((((((((((........((((((.....))))))(((((......))))))))((((....--.......))))..(((......))).)))))))........------ ( -30.70, z-score = -1.63, R) >droSim1.chr3R 2890184 104 + 27517382 CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGG------ .((((((((((........((((((.....))))))(((((......))))))))((((....--.......))))..(((......))).)))))))........------ ( -30.70, z-score = -1.63, R) >droSec1.super_6 2940918 103 + 4358794 CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGG-CAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGG------ .((((((((((........((((((.....))))))(((((......))))))))((-(....--.......)))...(((......))).)))))))........------ ( -30.00, z-score = -1.68, R) >droYak2.chr3R 21283350 104 - 28832112 CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGG------ .((((((((((........((((((.....))))))(((((......))))))))((((....--.......))))..(((......))).)))))))........------ ( -30.70, z-score = -1.63, R) >droEre2.scaffold_4770 3102522 104 + 17746568 CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGG------ .((((((((((........((((((.....))))))(((((......))))))))((((....--.......))))..(((......))).)))))))........------ ( -30.70, z-score = -1.63, R) >droAna3.scaffold_13340 18483347 104 - 23697760 CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGU------ .((((((((((........((((((.....))))))(((((......))))))))((((....--.......))))..(((......))).)))))))........------ ( -30.70, z-score = -1.65, R) >dp4.chr2 11321158 110 - 30794189 -GCAAUGAAGCAAUCAAUCAAUGCGAAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAGCGACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGC-GCAGUCCCGCAGU -(((((((..............(((((((((((((((((((......)))))((...))))))))).)))))))....(((......))).)))))))-((......))... ( -37.00, z-score = -2.72, R) >droPer1.super_3 5155470 110 - 7375914 -GCAAUGAAGCAAUCAAUCAAUGCGAAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAGCGACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGC-GCAGUCCCGCAGU -(((((((..............(((((((((((((((((((......)))))((...))))))))).)))))))....(((......))).)))))))-((......))... ( -37.00, z-score = -2.72, R) >droWil1.scaffold_181089 95119 101 - 12369635 -GCCAUAAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUUCAUCAUCAUUGCAGG-------- -........(((((..(((((((((.....)))))))))...........((...((((....--.......))))..))................)))))...-------- ( -22.20, z-score = -0.39, R) >droVir3.scaffold_13047 13393666 100 - 19223366 CGCAACGUUGCAAUCAAUCACAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUC---------- .......(((((((......((((((.....))...(((((......)))))))))(((....--.......)))...(((......)))...)))))))..---------- ( -24.90, z-score = -0.70, R) >droMoj3.scaffold_6540 17913749 100 - 34148556 CGCAAUGUUGCAAUCAAUCACUGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--AGUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUC---------- .((((((((((.....((((..(((.....)))..))))...))))....((...((((....--.......))))..))............))))))....---------- ( -23.90, z-score = 0.10, R) >droGri2.scaffold_14906 2986510 100 - 14172833 CGCAAUGAAGCAAUCAAUCACUGUGUAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG--ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUC---------- .((((((((((...........(((.....)))...(((((......))))))))((((....--.......))))..(((......))).)))))))....---------- ( -26.60, z-score = -1.51, R) >consensus CGCAAUGAAGCAAUCAAUCAAAGCGCAAAUCGCUUUGGUGUCGCAAAGCACCGCUGGGCAAAG__ACUUUUCGCCUUAGGAAUAAUAUCCAUCAUUGCAGUGGAGG______ .((((((((((...........(((.....)))...(((((......))))))))((((((((....)))).))))..(((......))).))))))).............. (-23.93 = -24.45 + 0.52)


| Location | 2,850,218 – 2,850,322 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.11 |
| Shannon entropy | 0.20854 |
| G+C content | 0.46845 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
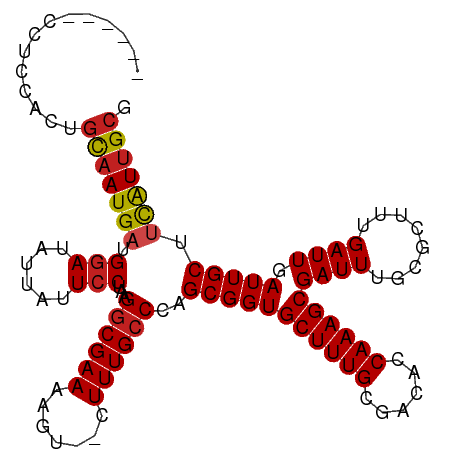
>dm3.chr3R 2850218 104 - 27905053 ------CCUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ------........(((((((.(((......)))...(((((..(((--(...((.(((...(((((((......)))))))..))).))...))))..)))))))))))). ( -31.30, z-score = -2.12, R) >droSim1.chr3R 2890184 104 - 27517382 ------CCUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ------........(((((((.(((......)))...(((((..(((--(...((.(((...(((((((......)))))))..))).))...))))..)))))))))))). ( -31.30, z-score = -2.12, R) >droSec1.super_6 2940918 103 - 4358794 ------CCUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUG-CCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ------........(((((((.(((......)))...((((((....--.))))-))(((((((........))))((((((.....))))))........)))))))))). ( -32.20, z-score = -2.36, R) >droYak2.chr3R 21283350 104 + 28832112 ------CCUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ------........(((((((.(((......)))...(((((..(((--(...((.(((...(((((((......)))))))..))).))...))))..)))))))))))). ( -31.30, z-score = -2.12, R) >droEre2.scaffold_4770 3102522 104 - 17746568 ------CCUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ------........(((((((.(((......)))...(((((..(((--(...((.(((...(((((((......)))))))..))).))...))))..)))))))))))). ( -31.30, z-score = -2.12, R) >droAna3.scaffold_13340 18483347 104 + 23697760 ------ACUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ------........(((((((.(((......)))...(((((..(((--(...((.(((...(((((((......)))))))..))).))...))))..)))))))))))). ( -31.30, z-score = -2.19, R) >dp4.chr2 11321158 110 + 30794189 ACUGCGGGACUGC-GCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGUCGCUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUUCGCAUUGAUUGAUUGCUUCAUUGC- ...(((....)))-(((((((.(((......)))....(((((((.((((((((....((((....)))).....)))))))))))))))..............)))))))- ( -35.70, z-score = -1.72, R) >droPer1.super_3 5155470 110 + 7375914 ACUGCGGGACUGC-GCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGUCGCUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUUCGCAUUGAUUGAUUGCUUCAUUGC- ...(((....)))-(((((((.(((......)))....(((((((.((((((((....((((....)))).....)))))))))))))))..............)))))))- ( -35.70, z-score = -1.72, R) >droWil1.scaffold_181089 95119 101 + 12369635 --------CCUGCAAUGAUGAUGAAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUUAUGGC- --------.(((((((((((.....))))))).....((((((....--.))))))..)))).(((..((((((.(((((((.....)))))))..)).)))).....)))- ( -25.40, z-score = -0.66, R) >droVir3.scaffold_13047 13393666 100 + 19223366 ----------GACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUGUGAUUGAUUGCAACGUUGCG ----------....((((((.((.(.((((.((....((((((....--.))))))..(((((((.((((.........))))...))))))))).))))).)).)))))). ( -30.70, z-score = -1.57, R) >droMoj3.scaffold_6540 17913749 100 + 34148556 ----------GACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAACU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCAGUGAUUGAUUGCAACAUUGCG ----------.(((((......(((......)))...((((((....--.))))))..)))))((((((......))))))......(((((((.(((....)))))))))) ( -31.50, z-score = -2.39, R) >droGri2.scaffold_14906 2986510 100 + 14172833 ----------GACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU--CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUACACAGUGAUUGAUUGCUUCAUUGCG ----------....(((((((.(((......)))...((((((....--.)))))).((((((((((((......))))))((((........)))).))))))))))))). ( -28.30, z-score = -1.83, R) >consensus ______CCUCCACUGCAAUGAUGGAUAUUAUUCCUAAGGCGAAAAGU__CUUUGCCCAGCGGUGCUUUGCGACACCAAAGCGAUUUGCGCUUUGAUUGAUUGCUUCAUUGCG ..............(((((((.(((......)))...((((((.......))))))..(((((((((((......))))))((((........)))).))))).))))))). (-24.43 = -24.70 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:13 2011