
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,837,111 – 2,837,216 |
| Length | 105 |
| Max. P | 0.965163 |

| Location | 2,837,111 – 2,837,214 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Shannon entropy | 0.47289 |
| G+C content | 0.44318 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.12 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
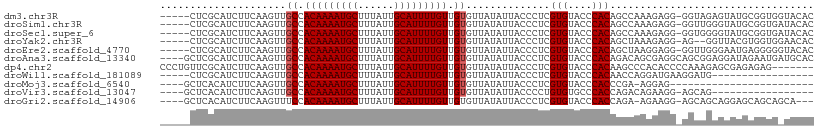
>dm3.chr3R 2837111 103 + 27905053 -----CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCAAAGAGG-GGUAGAGUAUGCGGUGGUACAC -----.((((((.(((.....((.(((((((((......))))))))).)).........(((((..((.......))....))))-)...))).))))))........ ( -29.40, z-score = -1.54, R) >droSim1.chr3R 2877293 103 + 27517382 -----CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCAAAGAGG-GGUUGGGUAUGCGGUGAUACAC -----...(((.(.....).))).(((((((((......))))))))).((((...((((((...(((((((((..(((.......-)))))))))))))))))))))) ( -33.10, z-score = -2.45, R) >droSec1.super_6 2927979 103 + 4358794 -----CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCAAAGAGG-GGUGGGGUAUGCGGUGAUACAC -----...(((.(.....).))).(((((((((......))))))))).((((...((((((...((((((((.((.((......)-).)))))))))))))))))))) ( -35.40, z-score = -2.77, R) >droYak2.chr3R 21260991 101 - 28832112 -----CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCUAAAGAGG-AG--GGUUACGUGGUGGAACAC -----...(((.(.....).))).(((((((((......))))))))).(((((....((((..((((.((((.(..(....)..)-.)--))))))).)))).))))) ( -29.20, z-score = -1.93, R) >droEre2.scaffold_4770 3090103 103 + 17746568 -----CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCUAAGGAGG-GGUUGGGAAUGAGGGGGUACAC -----.............((.((((((((((((......)))))))))............(((((((...(((((..((....)).-.).)))).)))))))))))).. ( -31.90, z-score = -1.61, R) >droAna3.scaffold_13340 18469433 105 - 23697760 ----GCUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGACAGCGAGGCAGCGGAGGAUAGAAUGAUGCAC ----....((((((((.....((.(((((((((......))))))))).))..........(((((((....))).....((......)).))))...))).))))).. ( -32.00, z-score = -2.26, R) >dp4.chr2 11299153 102 - 30794189 CCCUGUUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAAGCCCACACCCCAAAGAGCGAGAGAG------- ..((.(((((.((((......((.(((((((((......))))))))).))..............(((..(......)..)))......)))))))))))..------- ( -22.70, z-score = -3.09, R) >droWil1.scaffold_181089 12255995 87 + 12369635 -----CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAACCAGGAUGAAGGAUG----------------- -----......((((((....((.(((((((((......))))))))).))..........(((..(((....)))...))).))))))...----------------- ( -20.80, z-score = -2.00, R) >droMoj3.scaffold_6540 17898298 80 - 34148556 ----GCUCACAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACCCGA-AGGAG------------------------ ----.......(((((.....((.(((((((((......))))))))).))..............(((....)))..))-)))..------------------------ ( -17.70, z-score = -2.53, R) >droVir3.scaffold_13047 13371834 87 - 19223366 ----GCUCACAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCCUGUGUGCCCACCAGACAGAAGG-AGCAG----------------- ----((((.............((.(((((((((......))))))))).))............((((.((.....)).))))...)-)))..----------------- ( -24.10, z-score = -2.66, R) >droGri2.scaffold_14906 2970958 100 - 14172833 ----GCUCACAUCUUCAAGUUUCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACCAGA-AGAAGG-AGCAGCAGGAGCAGCAGCA--- ----((((.(.(((((........(((((((((......))))))))).(((.(((((.......)))))..)))..))-))).))-))).((....)).......--- ( -25.80, z-score = -1.88, R) >consensus _____CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCAAAGAGG_GGUAGGGUAUGCGGUG_UACAC .....................((.(((((((((......))))))))).))..............(((....))).................................. (-13.03 = -13.12 + 0.09)

| Location | 2,837,111 – 2,837,214 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Shannon entropy | 0.47289 |
| G+C content | 0.44318 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.64 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
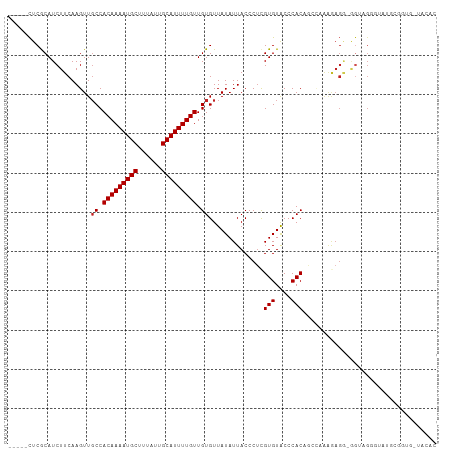
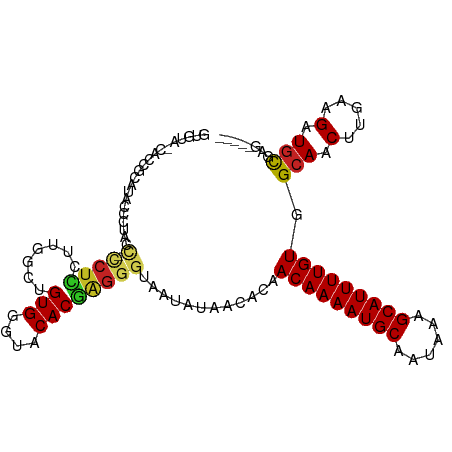
>dm3.chr3R 2837111 103 - 27905053 GUGUACCACCGCAUACUCUACC-CCUCUUUGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG----- (((((((..((..(((.((((.-((.....))..)))))))..))..)))......)))).(((((((((......))))))))).(((.(.....).)))...----- ( -28.30, z-score = -1.60, R) >droSim1.chr3R 2877293 103 - 27517382 GUGUAUCACCGCAUACCCAACC-CCUCUUUGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG----- (((((((..((..((((((...-((.....))...))))))..))..)))......)))).(((((((((......))))))))).(((.(.....).)))...----- ( -30.20, z-score = -2.09, R) >droSec1.super_6 2927979 103 - 4358794 GUGUAUCACCGCAUACCCCACC-CCUCUUUGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG----- (((((((..((..(((((((.(-(......)).)).)))))..))..)))......)))).(((((((((......))))))))).(((.(.....).)))...----- ( -29.20, z-score = -1.60, R) >droYak2.chr3R 21260991 101 + 28832112 GUGUUCCACCACGUAACC--CU-CCUCUUUAGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG----- (((((..(((.(((.(((--(.-(..(....)..).))))..)))..))).....))))).(((((((((......))))))))).(((.(.....).)))...----- ( -25.70, z-score = -1.26, R) >droEre2.scaffold_4770 3090103 103 - 17746568 GUGUACCCCCUCAUUCCCAACC-CCUCCUUAGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG----- ((((...(((((...((((...-.((....))...)))).....))))).......)))).(((((((((......))))))))).(((.(.....).)))...----- ( -26.80, z-score = -1.67, R) >droAna3.scaffold_13340 18469433 105 + 23697760 GUGCAUCAUUCUAUCCUCCGCUGCCUCGCUGUCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAGC---- .((((((.((((((((((...((((.(((.....)))))))...)))))))..........(((((((((......)))))))))(....)..)))))))))...---- ( -35.70, z-score = -3.34, R) >dp4.chr2 11299153 102 + 30794189 -------CUCUCUCGCUCUUUGGGGUGUGGGCUUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAACAGGG -------((((.((((((((..((.(((...((((((....))))))..............(((((((((......))))))))).))).))..)))).))))..)))) ( -36.00, z-score = -4.62, R) >droWil1.scaffold_181089 12255995 87 - 12369635 -----------------CAUCCUUCAUCCUGGUUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG----- -----------------((((.((((((((.((.((....)))).)))))...........(((((((((......)))))))))(....)..)))))))....----- ( -22.10, z-score = -1.67, R) >droMoj3.scaffold_6540 17898298 80 + 34148556 ------------------------CUCCU-UCGGGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGUGAGC---- ------------------------..(((-(((.((....)).))))))........(((((((((((((......)))))))))(....).......))))...---- ( -22.60, z-score = -2.83, R) >droVir3.scaffold_13047 13371834 87 + 19223366 -----------------CUGCU-CCUUCUGUCUGGUGGGCACACAGGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGUGAGC---- -----------------.((((-(((..((((.....))))...)))))))......(((((((((((((......)))))))))(....).......))))...---- ( -26.20, z-score = -2.16, R) >droGri2.scaffold_14906 2970958 100 + 14172833 ---UGCUGCUGCUCCUGCUGCU-CCUUCU-UCUGGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGAAACUUGAAGAUGUGAGC---- ---.((((((.(((.((.((((-(((...-...)).))))))).)))))).......(((((((((((((......)))))))))(....).......)))))))---- ( -27.40, z-score = -1.38, R) >consensus GUGUA_CACCGCAUACCCUACC_CCUCCUUGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG_____ .......................((((.......(((....))))))).............(((((((((......))))))))).(((.(.....).)))........ (-14.68 = -14.64 + -0.04)

| Location | 2,837,111 – 2,837,216 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Shannon entropy | 0.45281 |
| G+C content | 0.44449 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -13.63 |
| Energy contribution | -14.13 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
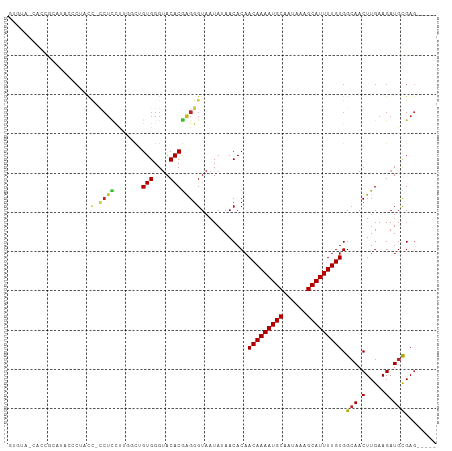
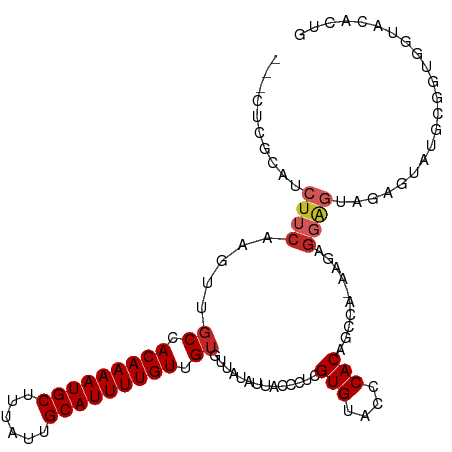
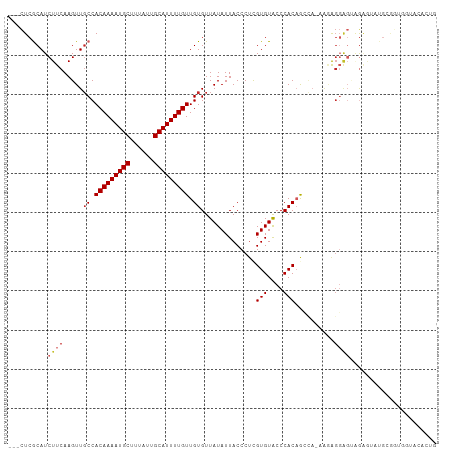
>dm3.chr3R 2837111 105 + 27905053 ---CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCA-AAGAGGGGUAGAGUAUGCGGUGGUACACUG ---............(((((((((...((((((((((.(.((((((((((.(((((.......)))))..))))).))-))).).))))))))))...)))))).))). ( -30.70, z-score = -1.71, R) >droSim1.chr3R 2877293 105 + 27517382 ---CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCA-AAGAGGGGUUGGGUAUGCGGUGAUACACUG ---...(((.(.....).))).(((((((((......))))))))).((((...((((((...(((((((((..(((.-......)))))))))))))))))))))).. ( -34.30, z-score = -2.62, R) >droSec1.super_6 2927979 105 + 4358794 ---CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCA-AAGAGGGGUGGGGUAUGCGGUGAUACACUG ---...(((.(.....).))).(((((((((......))))))))).((((...((((((...((((((((.((.((.-.....)).)))))))))))))))))))).. ( -36.60, z-score = -2.94, R) >droYak2.chr3R 21260991 103 - 28832112 ---CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCUA-AAGAGGAG--GGUUACGUGGUGGAACACUG ---...(((.(.....).))).(((((((((......))))))))).(((((....((((..((((.((((.(..(..-..)..).)--))))))).)))).))))).. ( -30.40, z-score = -2.14, R) >droEre2.scaffold_4770 3090103 105 + 17746568 ---CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCUA-AGGAGGGGUUGGGAAUGAGGGGGUACACUG ---............((((((((((((((((......)))))))))............(((((((...(((((..((.-...))..).)))).))))))))))).))). ( -33.50, z-score = -1.80, R) >droAna3.scaffold_13340 18469433 107 - 23697760 --GCUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGACAGCGAGGCAGCGGAGGAUAGAAUGAUGCACUG --....((((((((.....((.(((((((((......))))))))).))..........(((((((....))).....((......)).))))...))).))))).... ( -32.00, z-score = -1.98, R) >dp4.chr2 11299155 100 - 30794189 CUGUUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAAGCC-CACACCCC-AAAGAGCGAGAGAG------- ((.(((((.((((......((.(((((((((......))))))))).))..............(((..(......)..-))).....-.)))))))))))..------- ( -22.40, z-score = -2.68, R) >droWil1.scaffold_181089 12255995 87 + 12369635 ---CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAACC-----AGGAU-GAAGGAUG------------- ---......((((((....((.(((((((((......))))))))).))..........(((..(((....)))...-----))).)-)))))...------------- ( -20.80, z-score = -2.00, R) >droVir3.scaffold_13047 13371834 87 - 19223366 --GCUCACAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCCUGUGUGCCCACCAG-ACAGAAGGAGCAG------------------- --((((.............((.(((((((((......))))))))).))............((((.((.....)).-))))...))))..------------------- ( -24.10, z-score = -2.66, R) >droGri2.scaffold_14906 2970958 100 - 14172833 --GCUCACAUCUUCAAGUUUCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACCAG-A-AGAAGGAGCAGCAGGAGCAGCAGCA----- --((((.(.(((((........(((((((((......))))))))).(((.(((((.......)))))..)))..)-)-))).))))).((....)).......----- ( -25.80, z-score = -1.88, R) >consensus ___CUCGCAUCUUCAAGUUGCCACAAAAUGCUUUAUUGCAUUUUGUUGUGUUAUAUUACCCUCGUGUACCCACAGCCA_AAGAGGAGUAGAGUAUGCGGUGGUACACUG ..........((((.....((.(((((((((......))))))))).))..............(((....)))..........))))...................... (-13.63 = -14.13 + 0.50)

| Location | 2,837,111 – 2,837,216 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Shannon entropy | 0.45281 |
| G+C content | 0.44449 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
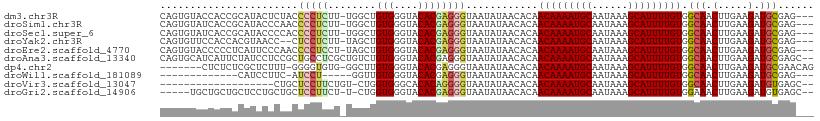
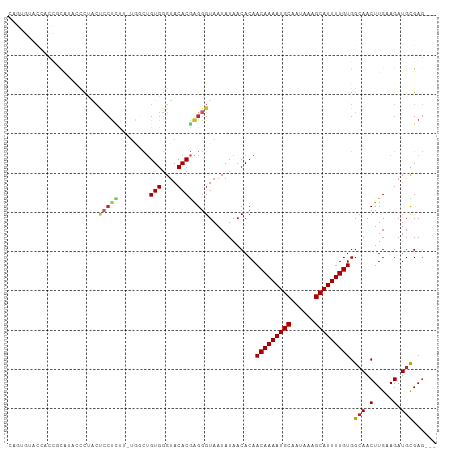
>dm3.chr3R 2837111 105 - 27905053 CAGUGUACCACCGCAUACUCUACCCCUCUU-UGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG--- ..(((((((..((..(((.((((.((....-.))..)))))))..))..)))......)))).(((((((((......))))))))).(((.(.....).)))...--- ( -28.50, z-score = -1.43, R) >droSim1.chr3R 2877293 105 - 27517382 CAGUGUAUCACCGCAUACCCAACCCCUCUU-UGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG--- ..(((((((..((..((((((...((....-.))...))))))..))..)))......)))).(((((((((......))))))))).(((.(.....).)))...--- ( -30.40, z-score = -1.98, R) >droSec1.super_6 2927979 105 - 4358794 CAGUGUAUCACCGCAUACCCCACCCCUCUU-UGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG--- ..(((((((..((..(((((((.((.....-.)).)).)))))..))..)))......)))).(((((((((......))))))))).(((.(.....).)))...--- ( -29.40, z-score = -1.52, R) >droYak2.chr3R 21260991 103 + 28832112 CAGUGUUCCACCACGUAACC--CUCCUCUU-UAGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG--- ..(((((..(((.(((.(((--(.(..(..-..)..).))))..)))..))).....))))).(((((((((......))))))))).(((.(.....).)))...--- ( -25.90, z-score = -1.17, R) >droEre2.scaffold_4770 3090103 105 - 17746568 CAGUGUACCCCCUCAUUCCCAACCCCUCCU-UAGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG--- ..((((...(((((...((((....((...-.))...)))).....))))).......)))).(((((((((......))))))))).(((.(.....).)))...--- ( -27.00, z-score = -1.53, R) >droAna3.scaffold_13340 18469433 107 + 23697760 CAGUGCAUCAUUCUAUCCUCCGCUGCCUCGCUGUCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAGC-- ...((((((.((((((((((...((((.(((.....)))))))...)))))))..........(((((((((......)))))))))(....)..)))))))))...-- ( -35.70, z-score = -3.08, R) >dp4.chr2 11299155 100 + 30794189 -------CUCUCUCGCUCUUU-GGGGUGUG-GGCUUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAACAG -------.....((((((((.-.((.(((.-..((((((....))))))..............(((((((((......))))))))).))).))..)))).)))).... ( -34.40, z-score = -4.26, R) >droWil1.scaffold_181089 12255995 87 - 12369635 -------------CAUCCUUC-AUCCU-----GGUUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG--- -------------((((.(((-(((((-----.((.((....)))).)))))...........(((((((((......)))))))))(....)..)))))))....--- ( -22.10, z-score = -1.67, R) >droVir3.scaffold_13047 13371834 87 + 19223366 -------------------CUGCUCCUUCUGU-CUGGUGGGCACACAGGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGUGAGC-- -------------------.(((((((..(((-(.....))))...)))))))......(((((((((((((......)))))))))(....).......))))...-- ( -26.20, z-score = -2.16, R) >droGri2.scaffold_14906 2970958 100 + 14172833 -----UGCUGCUGCUCCUGCUGCUCCUUCU-U-CUGGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGAAACUUGAAGAUGUGAGC-- -----.((((((.(((.((.(((((((...-.-..)).))))))).)))))).......(((((((((((((......)))))))))(....).......)))))))-- ( -27.40, z-score = -1.38, R) >consensus CAGUGUACCACCGCAUACCCUACUCCUCUU_UGGCUGUGGGUACACGAGGGUAAUAUAACACAACAAAAUGCAAUAAAGCAUUUUGUGGCAACUUGAAGAUGCGAG___ .......................(((((........(((....))))))))............(((((((((......))))))))).(((.(.....).)))...... (-15.61 = -15.75 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:11 2011