| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,785,178 – 2,785,274 |
| Length | 96 |
| Max. P | 0.555759 |

| Location | 2,785,178 – 2,785,274 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Shannon entropy | 0.45302 |
| G+C content | 0.56743 |
| Mean single sequence MFE | -35.41 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
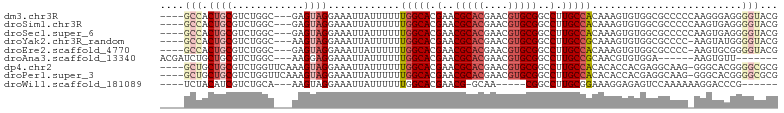
>dm3.chr3R 2785178 96 + 27905053 ----GCCACUGCGUCUGGC---GAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACAAAGUGUGGCGCCCCCAAGGGAGGGGUACG ----((((((((.((....---)))))).....((((((.(((((.(..(((((....)))))..).))))).))))))))))((((((....).)))))... ( -38.70, z-score = -1.67, R) >droSim1.chr3R 2824828 96 + 27517382 ----GCCACUGCGUCUGGC---GAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACAAAGUGUGGCGCCCCCAAGUGAGGGGUACG ----((((((((.((....---)))))).....((((((.(((((.(..(((((....)))))..).))))).))))))))))((((((....).)))))... ( -39.10, z-score = -2.03, R) >droSec1.super_6 2876328 96 + 4358794 ----GCCACUGCGUCUGGC---GAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACAAAGUGUGGCGCCCCCAAGUGAGGGGUACG ----((((((((.((....---)))))).....((((((.(((((.(..(((((....)))))..).))))).))))))))))((((((....).)))))... ( -39.10, z-score = -2.03, R) >droYak2.chr3R_random 662216 95 + 3277457 ----GCCACUGCGUCUGGC---AAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCGCAAAGUGUGGCGCCCC-AAGUAUGGGGUACG ----((((((((.....))---)...........(((((.(((((.(..(((((....)))))..).))))).))))))))))(((((-(....))))))... ( -39.80, z-score = -2.30, R) >droEre2.scaffold_4770 3036189 95 + 17746568 ----GCCACUGCGUCUGGC---GAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACAAAGUGUGGCGCCCC-AAGUGCGGGGUACG ----((((((((.((....---)))))).....((((((.(((((.(..(((((....)))))..).))))).))))))))))(((((-......)))))... ( -39.50, z-score = -1.81, R) >droAna3.scaffold_13340 18422913 87 - 23697760 ACGAUCUGCUGCGUCUGGC---AAGGAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCGCAACGUGUGGA------AAGUGUU------- ((..((..(.((((.((((---((((.(((((....)))))........(((((....))))).))))))))..)))))..))------..))...------- ( -30.30, z-score = -1.16, R) >dp4.chr2 11227154 98 - 30794189 ----GCUGCUGCGUCUGGUUCAAAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACACACCACGAGGCAAG-GGGCACGGGGCGCG ----......((((((.((..(((((((....)))))))...((.....(((((....))))).(((((((.(.......).))))))-).)))).)))))). ( -35.30, z-score = -1.07, R) >droPer1.super_3 5064409 98 - 7375914 ----GCUGCUGCGUCUGGUUCAAAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACACACCACGAGGCAAG-GGGCACGGGGCGCG ----......((((((.((..(((((((....)))))))...((.....(((((....))))).(((((((.(.......).))))))-).)))).)))))). ( -35.30, z-score = -1.07, R) >droWil1.scaffold_181089 12190820 84 + 12369635 ----UCUACAUCGUCUGCA---AAGUAGGAAAUUAUUUUUUGGCACGAACG-GCAA-----CGGCCUUGCGGAAAGGAGAGUCCAAAAAAGGACCCG------ ----(((.(....((((((---(.((..((((....))))..))......(-((..-----..))))))))))...))))((((......))))...------ ( -21.60, z-score = -0.93, R) >consensus ____GCCACUGCGUCUGGC___AAGUAGGAAAUUAUUUUUUGGCACGAACGCACGAACGUGCGGCCUUGCCACAAAGUGUGGCGCCCC_AAGUGCGGGGUACG ....(((.((((............))))............(((((.(..(((((....)))))..).))))).........................)))... (-17.59 = -18.48 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:01 2011