
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,782,196 – 2,782,295 |
| Length | 99 |
| Max. P | 0.638357 |

| Location | 2,782,196 – 2,782,295 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Shannon entropy | 0.50211 |
| G+C content | 0.35452 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -7.03 |
| Energy contribution | -7.22 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638357 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 2782196 99 + 27905053 UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUUCCCCCUCUUAUUGUUGCUUUGCGGCUGCUCUUGUUGUAAA--UGCAACAUCGAUGAAUGCGAAA ..........((((((.((((((((....)))))..............((((((((((((((.......))))))))--.)))))))))...))))))... ( -26.30, z-score = -3.25, R) >droAna3.scaffold_13340 18419769 97 - 23697760 UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUUUUUUCAGCCGCUGUCG--UUGCGGCUGCUCUUGUUGUAAC--UGCAACAUCGAUGAAUGCGAAA ..........((((((.(((.(((((.(((((((.......(((((((.....--..))))))).....))))))).--)))))..)))...))))))... ( -32.70, z-score = -3.68, R) >droEre2.scaffold_4770 3033164 99 + 17746568 UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUCCCCCCUCUUAUUGUUGUUUUGCGGCUGCUCUUGUUGUAAA--UGCAACAUCGAUGAAUGCGAAA ..........((((((.((((((((....)))))..............((((((((((((((.......))))))))--.)))))))))...))))))... ( -23.90, z-score = -2.59, R) >droYak2.chr3R_random 659322 99 + 3277457 UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUUCCCCUUCUUAUUGUUGUAUUGCGGCUGCUUUUGUUGUAAA--UACAACAUCGAUGAAUGCGAAA ..........((((((.((((((((....)))))..............((((((((((((((.......))))).))--))))))))))...))))))... ( -24.50, z-score = -2.91, R) >droSec1.super_6 2873371 99 + 4358794 UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUUCCCCCUCUUAUUGUUGUUUUGCGGCUGCUCUUGUUGUAAA--UGCAACAUCGAUGAAUGCGAAA ..........((((((.((((((((....)))))..............((((((((((((((.......))))))))--.)))))))))...))))))... ( -23.90, z-score = -2.46, R) >droSim1.chr3R 2820672 99 + 27517382 UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUCCCCCCUCUUAUUGUUGUUUUGCGGCUGCUCUUGUUGUAAA--UGCAACAUCGAUGAAUGCGAAA ..........((((((.((((((((....)))))..............((((((((((((((.......))))))))--.)))))))))...))))))... ( -23.90, z-score = -2.59, R) >droVir3.scaffold_13047 13301184 88 - 19223366 UUUAAAUUUAUGCCCUACGAAUUGCAUUUACAAUUUUUUCUUUA-UUUCCCCC----------GCCGUUGUUGUAAA--UGCAUUUCCAUUCAAUGCAAAA ..........(((.....(((.((((((((((((..........-........----------......))))))))--)))).)))........)))... ( -15.68, z-score = -3.77, R) >droMoj3.scaffold_6540 16319262 98 + 34148556 UUUAAAUUUAUGCCCUACGAAUUGCAUUUACAAUUUUUUGCUCA-UGUUGCCCUGCAUCAGAUGUGGUUGUUGUAAA--UGCAUUGCCAUUGAAUGCAAAA ......................((((((((((((...(..(...-((.(((...))).))...)..)..))))))))--))))((((........)))).. ( -22.50, z-score = -1.27, R) >droGri2.scaffold_14906 2899435 98 - 14172833 UUUAAAUUUAUGCCCUACGAAUUGCAUUUACAAUUUUUUA--UG-UGUUGUUCUCGUUUCGGUUUUUGUGCUGCAACGUUGCAUUGCCGAUGAAUGCAAAA .....................(((((((((((((......--..-.))))........(((((....((((.((...)).)))).)))))))))))))).. ( -19.20, z-score = -0.46, R) >droWil1.scaffold_181089 12187471 99 + 12369635 UUUAAAUUUAUGCGCUACGAAUUGCAUUUGCAAUGUUUUUUGCAACCUUUUUUCCUUUUGCUCUUUGUAGUUGCAAA--UGCAACAUCACUGAAUGCAAAA ..........((((....((.(((((((((((((.......((((............))))........))))))))--)))))..))......))))... ( -20.96, z-score = -1.27, R) >consensus UUUAAAUUUAUGCGUUACGAAUUGCAUUUGCAAUUUUCCCCCUCUUAUUGUUGUUUUGCGGCUGCUCUUGUUGUAAA__UGCAACAUCGAUGAAUGCGAAA .....................(((((((((((((............)))))....................((((....))))........)))))))).. ( -7.03 = -7.22 + 0.19)
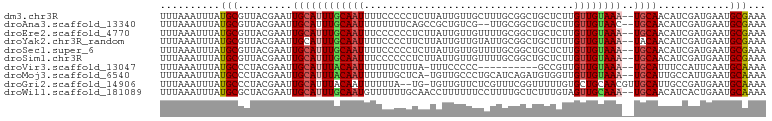

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:00 2011