
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,764,801 – 2,764,902 |
| Length | 101 |
| Max. P | 0.530211 |

| Location | 2,764,801 – 2,764,902 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Shannon entropy | 0.31622 |
| G+C content | 0.40080 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
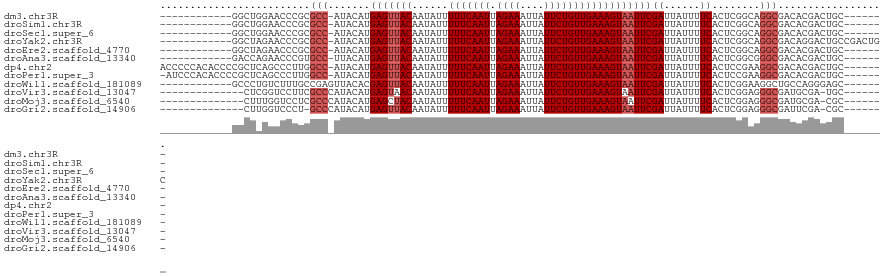
>dm3.chr3R 2764801 101 + 27905053 ------------GGCUGGAACCCGCGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGCAGGCGACACGACUGC------- ------------((((((...))).)))-.....((((((((......(((((((.((((....)))))))))))))))))))..............((((.((...)).))))------- ( -21.40, z-score = -1.01, R) >droSim1.chr3R 2803220 101 + 27517382 ------------GGCUGGAACCCGCGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGCAGGCGACACGACUGC------- ------------((((((...))).)))-.....((((((((......(((((((.((((....)))))))))))))))))))..............((((.((...)).))))------- ( -21.40, z-score = -1.01, R) >droSec1.super_6 2855953 101 + 4358794 ------------GGCUGGAACCCGCGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGCAGGCGACACGACUGC------- ------------((((((...))).)))-.....((((((((......(((((((.((((....)))))))))))))))))))..............((((.((...)).))))------- ( -21.40, z-score = -1.01, R) >droYak2.chr3R 7634348 108 + 28832112 ------------GGCUAGAACCCGCGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGCAGGCGACAGGACUGCCGACUGC ------------(((..........)))-.....((((((((......(((((((.((((....)))))))))))))))))))........((.(((((((.........))))))).)). ( -27.70, z-score = -2.32, R) >droEre2.scaffold_4770 3016068 101 + 17746568 ------------GGCUAGAACCCGCGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGCAGGCGACACGACUGC------- ------------.((.((......((((-......(((((((......(((((((.((((....))))))))))))))))))((......))........))))......))))------- ( -20.20, z-score = -0.98, R) >droAna3.scaffold_13340 18403649 101 - 23697760 ------------GACCAGAACCCGUGCC-UUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGCGGGCGACACGACUGC------- ------------...(((......((((-(..(..(((((((......(((((((.((((....))))))))))))))))))((......))....)..)))))......))).------- ( -20.70, z-score = -1.13, R) >dp4.chr2 11197898 113 - 30794189 ACCCCCACACCCCGCUCAGCCCUUGGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCCGAAGGCGACACGACUGC------- ............((....(((.((((..-......(((((((......(((((((.((((....))))))))))))))))))((......))...)))).)))....)).....------- ( -21.70, z-score = -2.07, R) >droPer1.super_3 5035861 112 - 7375914 -AUCCCACACCCCGCUCAGCCCUUGGCC-AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCCGAAGGCGACACGACUGC------- -...........((....(((.((((..-......(((((((......(((((((.((((....))))))))))))))))))((......))...)))).)))....)).....------- ( -21.70, z-score = -1.89, R) >droWil1.scaffold_181089 12166804 102 + 12369635 ------------GCCCUGUCUUUGCCGAGUUACACGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGAAGGCUGCCAGGGAGC------- ------------.(((((.((((.((((((.....(((((((......(((((((.((((....))))))))))))))))))((......))))))))))))....)))))...------- ( -30.60, z-score = -3.48, R) >droVir3.scaffold_13047 13278827 99 - 19223366 --------------CUCGGUCCUUCGCCCAUACAUGAGUAACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGAGGGCGAUGCGA-UGC------- --------------....((((.((((((.....(((((((((..((.((((.....))))..))..))))(((((((((...))))))))))))))..)))))).).))-)..------- ( -25.60, z-score = -2.27, R) >droMoj3.scaffold_6540 17806274 99 - 34148556 --------------CUUUGGUCCUCGCCCAUACAUGAGCUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGAGGGCGAUGCGA-CGC------- --------------.....((((((((((...................((((.....))))....((((.((((((((((...))))))))))..)))))))))).).))-)..------- ( -26.30, z-score = -2.55, R) >droGri2.scaffold_14906 2878043 98 - 14172833 --------------CUUGGUCCCU-GCCCAUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGAGGGCGAUUCGA-CGC------- --------------....(((..(-((((......(((((((......(((((((.((((....))))))))))))))))))((......)).......)))))....))-)..------- ( -22.90, z-score = -1.78, R) >consensus ____________GGCUAGAACCCGCGCC_AUACAUGAGUUACAAUAUUUUUCAAUUAGAAAUUAUUCUGUUGAAAGUAAUUCGAUUAUUUUCACUCGGAAGGCGACACGACUGC_______ .........................(((.......(((((((......(((((((.((((....))))))))))))))))))((......))........))).................. (-14.34 = -14.76 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:59 2011