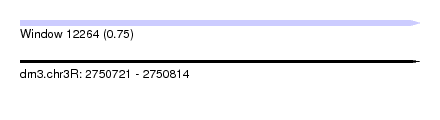
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,750,721 – 2,750,814 |
| Length | 93 |
| Max. P | 0.748781 |

| Location | 2,750,721 – 2,750,814 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.53 |
| Shannon entropy | 0.66362 |
| G+C content | 0.55526 |
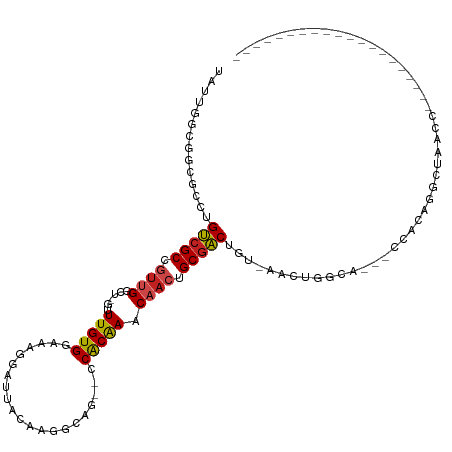
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -12.07 |
| Energy contribution | -11.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
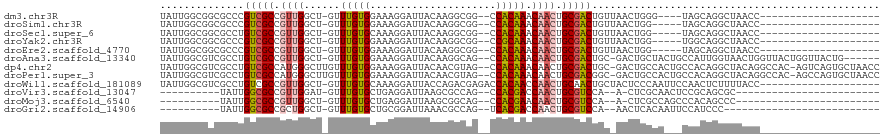
>dm3.chr3R 2750721 93 + 27905053 UAUUGGCGGCGCCCGUCGCCGUUGGCU-GUUUGUGGAAAGGAUUACAAGGCGG--CCACAAACAACUGCGACUGUUAACUGGG----UAGCAGGCUAACC-------------------- ....((((((....))))))(((((((-((((((((...(..........)..--))))))))).((((.(((........))----).)))))))))).-------------------- ( -36.80, z-score = -1.90, R) >droSim1.chr3R 2789075 92 + 27517382 UAUUGGCGGCGCCCGUCGCCGUUGGCU-GUUUGUGGAAAGGAUUACAAGGCGG--CCACAAACAACUGCGACUGUUAACUGG-----UAGCAGGCUAACC-------------------- ....((((((....))))))(((((((-((((((((...(..........)..--))))))))).((((.(((.......))-----).)))))))))).-------------------- ( -36.90, z-score = -2.16, R) >droSec1.super_6 2842045 92 + 4358794 UAUUGGCGGCGCCUGUCGCCGUUGGCU-GUUUGUGCAAAGGAUUACAAGGCGG--CCACAAACAACUGCGACUGUUAACUGG-----UAGCAGGCUAACC-------------------- ....((((((....))))))(((((((-((((((((...(..........).)--.)))))))).((((.(((.......))-----).)))))))))).-------------------- ( -31.90, z-score = -0.60, R) >droYak2.chr3R 21171718 92 - 28832112 UAUUGGCGGCGCCCGUCGCCGUUGGCU-GUUUGUGGAAAGGAUUACAAGGCGG--CCGCAAACAACUGCGACUGUUAACUGG-----UGGCAGGCUAACC-------------------- ....((((((....))))))(((((((-((((((((...(..........)..--))))))))).((((.(((.......))-----).)))))))))).-------------------- ( -38.60, z-score = -2.06, R) >droEre2.scaffold_4770 3002165 92 + 17746568 UAUUGGCGGCGCCCGUCGCCGUUGGCU-GUUUGUGGAAAGGAUUACAAGGCGG--CCACAAACAACUGCGACUGUUAACUGG-----UAGCAGGCUAACC-------------------- ....((((((....))))))(((((((-((((((((...(..........)..--))))))))).((((.(((.......))-----).)))))))))).-------------------- ( -36.90, z-score = -2.16, R) >droAna3.scaffold_13340 18390511 110 - 23697760 UAUUGGCGUCGCCUGUCGCCGUUGGCU-GUUUGUGGAAAGGAUUACAAGGCAG--CCACAAACAACUGCGACUGC-GACUGCUACUGCCAUUGGUAACUGGUUACUGGUUACUG------ ...(((((((((..(((((.(((((((-((((((((......)))))).))))--)))......)).))))).))-))).))))........((((((..(...)..)))))).------ ( -42.20, z-score = -2.90, R) >dp4.chr2 11179570 116 - 30794189 UAUUGGCGUCGCCUGUCGCCAUGGGCUUGUUUGUGGAAAGGAUUACAACGUAG--CCACAAACAACUGCGACUGC-GACUGCCACUGCCACAGGCUACAGGCCAC-AGUCAGUGCUAACC (((((((((.((((((.(((.(((((((((((((((...(........)....--)))))))))).(((....))-).........))).))))).)))))).))-.)))))))...... ( -44.50, z-score = -2.21, R) >droPer1.super_3 5017397 116 - 7375914 UAUUGGCGUCGCCUGUCGCCAUGGGCUUGUUUGUGGAAAGGAUUACAACGUAG--CCACAAACAACUGCGACGGC-GACUGCCACUGCCACAGGCUACAGGCCAC-AGCCAGUGCUAACC (((((((((.((((((.(((.(((((((((((((((...(........)....--)))))))))).......(((-....)))...))).))))).)))))).))-.)))))))...... ( -50.70, z-score = -3.68, R) >droWil1.scaffold_181089 12151083 99 + 12369635 UAUUGGCGUCGCCUGUCGCCGUUGGCU-GUUUGUGCAAAGGAUUACCAGACGAGACCACAACCAACUGCAACUGCUACUCCCAAUUCCAACUCUUUUACC-------------------- .(((((.((.((..((.((.(((((.(-((((((.(...((....)).)))))....))).))))).)).)).)).))..)))))...............-------------------- ( -19.30, z-score = 0.29, R) >droVir3.scaffold_13047 13262087 80 - 19223366 ----------UAUUGGCGCCGUUGGAU-GUUUGUGCUGAGGAUUAAGCGCCAG--CCACGACCAACUGCGUCCA--A-CUCGCAACUCCGCAGCGC------------------------ ----------.....((((.(((((((-((((((((((..(......)..)))--.)))))......)))))))--)-)..((......)).))))------------------------ ( -25.30, z-score = -0.24, R) >droMoj3.scaffold_6540 17789039 80 - 34148556 ----------UAUUGGCGCCGUUGGCU-GUUUGUGCUGAGGAUUAAGCGGCAG--CCACGAACAACUGCGUCCA--A-CUCGCCAGCCCACAGCCC------------------------ ----------..((((((..(((((((-((((((((((..(......)..)))--.)))))))).....).)))--)-).))))))..........------------------------ ( -25.60, z-score = -0.07, R) >droGri2.scaffold_14906 2862211 79 - 14172833 ----------UAUUGGCGCCGCUGGCU-GUUUGUGCUGCGGAUUAAACGCCAG--UCACGACCAACUGCGUCCA--AACUCACAAUUCCAUCCC-------------------------- ----------..(((((((.(.(((..-..((((((((((.......)).)))--.)))))))).).))).)))--).................-------------------------- ( -21.40, z-score = -1.13, R) >consensus UAUUGGCGGCGCCUGUCGCCGUUGGCU_GUUUGUGGAAAGGAUUACAAGGCAG__CCACAAACAACUGCGACUGU_AACUGGCA___CCACAGGCUAACC____________________ ..............(((((.((((......(((((.....................))))).)))).)))))................................................ (-12.07 = -11.93 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:58 2011