
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,736,769 – 2,736,883 |
| Length | 114 |
| Max. P | 0.884366 |

| Location | 2,736,769 – 2,736,883 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Shannon entropy | 0.26949 |
| G+C content | 0.43133 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.77 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
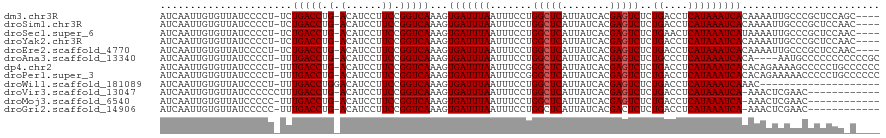
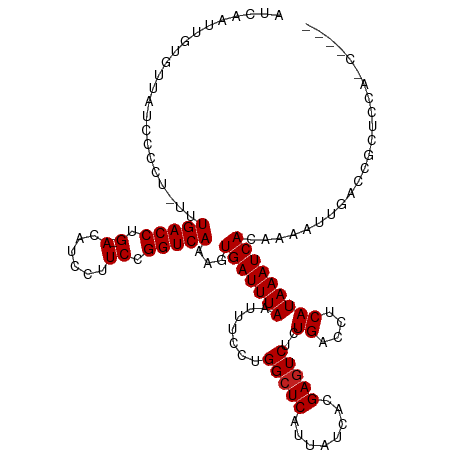
>dm3.chr3R 2736769 114 + 27905053 AUCAAUUGUGUUAUCCCCU-UCUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACAAAAUUGCCCGCUCCAGC---- ..(((((............-..(((((.(-(......)).)))))..((((((((...((..(((((........)))))...)).....))))))))..)))))...........---- ( -21.90, z-score = -1.98, R) >droSim1.chr3R 2773482 114 + 27517382 AUCAAUUGUGUUAUCCCCU-UCUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACAAAAUUGCCCGCUCCAAC---- ..(((((............-..(((((.(-(......)).)))))..((((((((...((..(((((........)))))...)).....))))))))..)))))...........---- ( -21.90, z-score = -2.40, R) >droSec1.super_6 2828295 114 + 4358794 AUCAAUUGUGUUAUCCCCU-UCUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGAACUCAUAAAUCAUAAAAUUGCCCGCUCCAAC---- ..(((((............-..(((((.(-(......)).)))))..((((((((..(((..(((((........)))))...)))....))))))))..)))))...........---- ( -20.90, z-score = -2.12, R) >droYak2.chr3R 21157541 114 - 28832112 AUCAAUUGUGUUAUCCCCU-UCUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACAAAAUUGCCCGCUCCAAC---- ..(((((............-..(((((.(-(......)).)))))..((((((((...((..(((((........)))))...)).....))))))))..)))))...........---- ( -21.90, z-score = -2.40, R) >droEre2.scaffold_4770 2987967 114 + 17746568 AUCAAUUGUGUUAUCCCCU-UCUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACAAAAUUGCCCGCUCCAAC---- ..(((((............-..(((((.(-(......)).)))))..((((((((...((..(((((........)))))...)).....))))))))..)))))...........---- ( -21.90, z-score = -2.40, R) >droAna3.scaffold_13340 18377570 114 - 23697760 AUCAAUUGUGUUAUCCCCU-UUUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGCCCUCAUAAAUCACA----AAUGCCCCCCCCCCCGC .......(((((.......-(((((((.(-(......)).)))))))((((((((.......(((((........)))))..........)))))))).----)))))............ ( -22.93, z-score = -3.55, R) >dp4.chr2 11162669 118 - 30794189 AUCAAUUGUGUUAUCCCCU-UUUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCGGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACACAGAAAAGCCCCCUGCCCCCC .....((.(((........-(((((((.(-(......)).)))))))((((((((...((.((((((........))))).).)).....))))))))))).))................ ( -24.60, z-score = -2.16, R) >droPer1.super_3 5000030 118 - 7375914 AUCAAUUGUGUUAUCCCCU-UUUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCGGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACACAGAAAAACCCCCUGCCCCCC .....((.(((........-(((((((.(-(......)).)))))))((((((((...((.((((((........))))).).)).....))))))))))).))................ ( -24.60, z-score = -2.50, R) >droWil1.scaffold_181089 12135508 99 + 12369635 AUCAAUUGUGUUAUCCCCU-UUUGACCUGGACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCAAAC-------------------- ..................(-(((((((.(((......)))))))))))(((((((...((..(((((........)))))...)).....)))))))...-------------------- ( -21.40, z-score = -2.13, R) >droVir3.scaffold_13047 13245668 106 - 19223366 AUCAAUUGUGUUAUCCCCCCUUUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCA-AAACUCGAAC------------ .....(((.(((.......((((((((.(-(......)).))))))))(((((((...((..(((((........)))))...)).....)))))))-.))).)))..------------ ( -23.90, z-score = -3.36, R) >droMoj3.scaffold_6540 17772040 105 - 34148556 AUCAAUUGUGUUAUCCCCC-UUUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCA-AAACUCGAAC------------ .....(((.(((......(-(((((((.(-(......)).))))))))(((((((...((..(((((........)))))...)).....)))))))-.))).)))..------------ ( -23.90, z-score = -3.34, R) >droGri2.scaffold_14906 2846761 105 - 14172833 AUCAAUUGUGUUAUCCCCC-UUUGACCUG-ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGACUCUCUGACCUCAUAAAUCA-AAACUCGAAC------------ .....(((.(((......(-(((((((.(-(......)).))))))))(((((((.......((.(((..............)))))...)))))))-.))).)))..------------ ( -18.54, z-score = -1.89, R) >consensus AUCAAUUGUGUUAUCCCCU_UUUGACCUG_ACAUCCUUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACAAAAUUGACCGCUCCA_C____ ......................(((((.............)))))...(((((((.......(((((........)))))..((....)))))))))....................... (-13.69 = -13.77 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:56 2011