
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,693,865 – 2,693,958 |
| Length | 93 |
| Max. P | 0.727020 |

| Location | 2,693,865 – 2,693,958 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Shannon entropy | 0.39015 |
| G+C content | 0.46719 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -14.61 |
| Energy contribution | -14.65 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
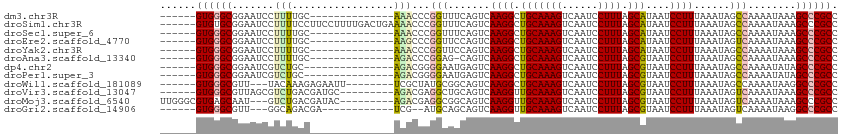
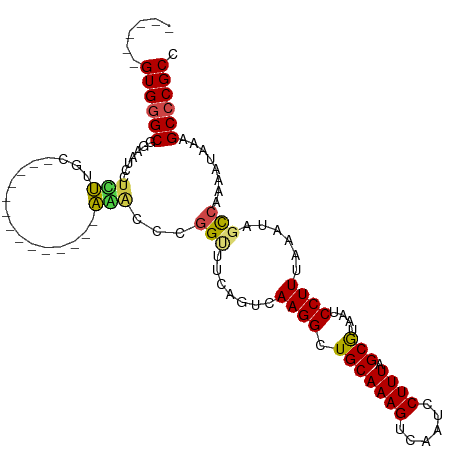
>dm3.chr3R 2693865 93 + 27905053 ------GUGGGCGGAAUCCUUUUGC--------------AAACCCGGUUUCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCAUAAUCCUUUAAAUAGCCAAAAUAAAGCCCGCC ------((((((((((((.......--------------......))))))......((((((((((......)))).)).(((....)))...))))........)))))). ( -22.42, z-score = -1.32, R) >droSim1.chr3R 2728432 107 + 27517382 ------GUGUGCGGAAUCCUUUUCCUUCCUUUUGACUGAAAACCCGGUUUCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCAUAAUCCUUUAAAUAGCCAAAAUAAAGCCCGCC ------(((.((((((.....))))......((((((((((......))))))))))((((((((((......)))).)).(((....)))...))))........)).))). ( -27.20, z-score = -3.36, R) >droSec1.super_6 2785121 93 + 4358794 ------GUGGGCGGAAUCCUUUUGC--------------AAACCCGGUUUCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCAUAAUCCUUUAAAUAGCCAAAAUAAAGCCCGCC ------((((((((((((.......--------------......))))))......((((((((((......)))).)).(((....)))...))))........)))))). ( -22.42, z-score = -1.32, R) >droEre2.scaffold_4770 2944334 93 + 17746568 ------GUGGGCGGAAUCCUUUUGC--------------AAGCCCGGUUCCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCAUAAUCCUUUAAAUAGUCAAAAUAAAGCCCGCC ------(((((((((....((((((--------------(.(((..(.......)..))))))))))....)))............((......))..........)))))). ( -24.30, z-score = -2.06, R) >droYak2.chr3R 21113667 93 - 28832112 ------GUGGGCGGAAUCCUUUUGC--------------AAACCCGGUUCCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCAUAAUCCUUUAAAUAGCCAAAAUAAAGCCCGCC ------((((((((((((.......--------------......))))))......((((((((((......)))).)).(((....)))...))))........)))))). ( -25.32, z-score = -2.42, R) >droAna3.scaffold_13340 18335584 92 - 23697760 ------GUGGGCGGAAUCCUUUUGC--------------AGACCCGGAG-CAGUCAAGGCUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGCCAAAAUAAAGCCCGCC ------(((((((((....((((((--------------((.((..((.-...))..))))))))))....)))(((.((.(((....)))....)).))).....)))))). ( -27.70, z-score = -2.12, R) >dp4.chr2 11117774 92 - 30794189 ------GUGGGCGGAAUCGUCUGC---------------AGACGGGGAAUGAGUCAAGGCUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGCCAAAAUAUAGCCCGCC ------((((((....(((((...---------------.)))))............((((((((((......)))).)).(((....)))...))))........)))))). ( -24.80, z-score = -0.63, R) >droPer1.super_3 4956066 92 - 7375914 ------GUGGGCGGAAUCGUCUGC---------------AGACGGGGAAUGAGUCAAGGCUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGCCAAAAUAUAGCCCGCC ------((((((....(((((...---------------.)))))............((((((((((......)))).)).(((....)))...))))........)))))). ( -24.80, z-score = -0.63, R) >droWil1.scaffold_181089 297284 96 - 12369635 ------GUGGGCGUU---UACAAAGAGAAUU--------UCGCUAUGCGGCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGCCAAAAUAAGGCCCGCC ------(((((((..---..)..((((.(((--------.(((((((((((.......))))))(((......)))))))).))).))))................)))))). ( -26.30, z-score = -1.43, R) >droVir3.scaffold_13047 6037812 98 + 19223366 ------GUGGGCGUUAGCGUCUGACGAUGC---------AGACGAGGCUGCAGUCAAGGUUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGUCAAAAUAAAGCCCGCC ------(((((((((.(((((....)))))---------.)))..(((((...(.((((.(((((((......)))).)))....)))).)..)))))........)))))). ( -31.80, z-score = -2.95, R) >droMoj3.scaffold_6540 16438466 101 + 34148556 UUGGGCGUGAGCAAU---GUCUGACGAUAC---------AGACGAGGCGGCAGUCAAGGUUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGUCAAAAUAAAGCCCGCC ..((((....((..(---(((((......)---------)))))..))(((.((.((((.(((((((......)))).)))....))))...)).)))........))))... ( -27.70, z-score = -1.63, R) >droGri2.scaffold_14906 11383781 90 + 14172833 ------GUGGGCGUU---GGCAGACGA------------UCG--AUGCAGCAGUCAAGGUUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGUCAAAAUAAGGCCCGCC ------((((((.((---(...(((..------------..(--((......)))((((.(((((((......)))).)))....))))......)))....))).)))))). ( -24.60, z-score = -1.60, R) >consensus ______GUGGGCGGAAUCCUCUUGC______________AAACCCGGUUUCAGUCAAGGCUGCAAAGUCAAUCCUUUAGCGUAAUCCUUUAAAUAGCCAAAAUAAAGCCCGCC ......((((((.................................(((....)))..((((((((((......)))).)).(((....)))...))))........)))))). (-14.61 = -14.65 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:48 2011