
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,687,637 – 2,687,743 |
| Length | 106 |
| Max. P | 0.886239 |

| Location | 2,687,637 – 2,687,743 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.42727 |
| G+C content | 0.44644 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2687637 106 - 27905053 AGAACUCGUGCGAUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGCCCGACUAGCUGCA-UGUCCCAUGUCCCUUAUAUCCUU ......((((((..(((((((((((..(((....)))..))))))))-)))(((((..((......))..))))).....))))-))..................... ( -25.30, z-score = -2.64, R) >droSim1.chr3R 2722220 106 - 27517382 AGAACUCGUGCGAUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGCCCGACUAGCUGCA-UGUCCCAUGUCCCUUAUAUCCUU ......((((((..(((((((((((..(((....)))..))))))))-)))(((((..((......))..))))).....))))-))..................... ( -25.30, z-score = -2.64, R) >droSec1.super_6 2778933 106 - 4358794 AGAACUCGUGCGAUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGCCCGACUAGCUGCA-UGUCCCUUGUCCCUUAUAUCCUU ......((((((..(((((((((((..(((....)))..))))))))-)))(((((..((......))..))))).....))))-))..................... ( -25.30, z-score = -2.87, R) >droYak2.chr3R 21107334 106 + 28832112 CGAACUCGUGCGUUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGCCCGACUAGCUGCA-UUUCCCUUGUCCUUUAUAUCCUU .(((...((((...(((((((((((..(((....)))..))))))))-)))(((((..((......))..)))))......)))-))))................... ( -23.30, z-score = -2.51, R) >droEre2.scaffold_4770 2937942 106 - 17746568 AGAACUCGUGCGUUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGCCCGACUAGCUGCA-UGUCCCUUCUCCCUUAUAUCCUU ((((..(((((...(((((((((((..(((....)))..))))))))-)))(((((..((......))..)))))......)))-))....))))............. ( -26.50, z-score = -3.65, R) >droAna3.scaffold_13340 18330125 102 + 23697760 AGAAUUCGUGCGUUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGAACAUCCCAUUGUGCCUGAGUAGCUGGA-UGUCCGUUUCUCUCCUCCU---- ((((..((..(((((((((((((((..(((....)))..))))))))-)))(((((.(((......))).))))).......))-))..)).))))........---- ( -25.00, z-score = -1.80, R) >droPer1.super_3 4950429 104 + 7375914 AGAAUUCGUGCGUUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGUCGGAGUGGCUGCAGUAUGUCCCGGUCCCUGUGCC--- .......(..(...(((((((((((..(((....)))..))))))))-)))..((((((.(((.........)))(.(((........))).))))))).)..).--- ( -28.60, z-score = -1.38, R) >dp4.chr2 11112207 104 + 30794189 AGAAUUCGUGCGUUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGUCGGAGUGGCUGCAGUAUGUCCCGGUCCCUGCGCC--- .......((((...(((((((((((..(((....)))..))))))))-)))..((((((.(((.........)))(.(((........))).))))))).)))).--- ( -31.00, z-score = -1.89, R) >droMoj3.scaffold_6540 16431673 106 - 34148556 GCGUUUCGUGCGUUUUUAUUACCGAAUUAAUAAAUUACCUUGACAGUGGAAUCGGGUCCAUCCCAUUGUGCGAGCGU-GCCGCAUCGUCUACCCCAUCCUUCACUCU- ..((..((((((..(((((((......))))))).((((((((((((((.((.(....))).))))))).)))).))-).)))).))...))...............- ( -21.80, z-score = -0.34, R) >droGri2.scaffold_14906 11377714 89 - 14172833 ACGAUUCGUGCGUUUUUAUUGCCGAAUUAAUAAAUUACCUUGACAGU-GAAUCGGGACCAUCCCAUUGUGUGUGUGC-CGUACCCCAUCCA----------------- ..(((..(((((..(((((((.(((..(((....)))..))).))))-)))..(((.....))).............-)))))...)))..----------------- ( -15.60, z-score = 0.51, R) >consensus AGAACUCGUGCGUUUUUAUUGUCGAAUUAAUAAAUUACCUUGACAGU_GAAUCGGGACCAUCCCAUUGUGCCCGACUAGCUGCA_UGUCCCUUGUCCCUUAUAUCCU_ .......(..((.(((.((((((((..(((....)))..)))))))).))).)(((.....)))...)..)..................................... (-13.76 = -13.85 + 0.09)

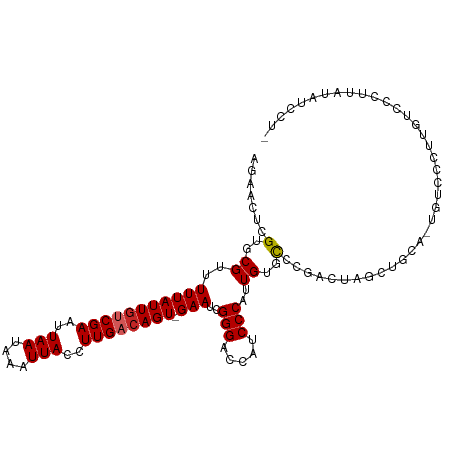
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:48 2011