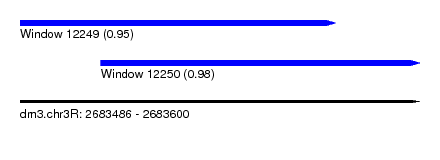
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,683,486 – 2,683,600 |
| Length | 114 |
| Max. P | 0.981933 |

| Location | 2,683,486 – 2,683,576 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Shannon entropy | 0.45340 |
| G+C content | 0.45327 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.04 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
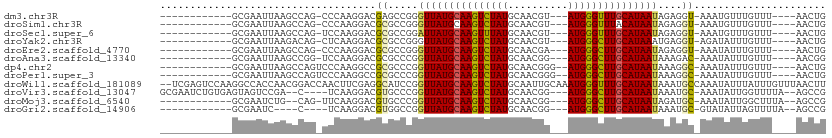
>dm3.chr3R 2683486 90 + 27905053 ------------GCGAAUUAAGCCAG-CCCAAGGACGAGCCGGGUUAUGCAAGUCUAUGCAACGU---AUGGGUUUGCAUAAUAGAGGU-AAAUGUUUGUUU----AACUG ------------((((((...(((.(-(((..((.....))))))(((((((..((((((...))---))))..))))))).....)))-....))))))..----..... ( -27.50, z-score = -2.15, R) >droSim1.chr3R 2718224 90 + 27517382 ------------GCGAAUUAAGCCAG-CCCAAGGACGCGCCGGGUUAUGCAAGUCUAUGCAACGU---AUGGGUUUACAUAAUAGAGGU-AAAUGUUUGUUU----AACUG ------------((((((...(((.(-(((..((.....))))))((((.((..((((((...))---))))..)).)))).....)))-....))))))..----..... ( -20.90, z-score = 0.17, R) >droSec1.super_6 2774929 90 + 4358794 ------------GCGAAUUAAGCCAG-UCCAAGGACGCGCCGGAUUAUGCAAGUUUAUGCAACGU---AUGGGUUUGCAUAAUAGAGGU-AAAUGUUUGUUU----AACUG ------------((((((...((..(-((....)))))(((..((((((((((..(((((...))---)))..))))))))))...)))-....))))))..----..... ( -26.70, z-score = -2.23, R) >droYak2.chr3R 21102822 90 - 28832112 ------------GCGAAUUAAGACAG-CUCAAGGACGCGCCGGGUUAUGCAAGUCUAUGCAACGU---AUGGGCUUGCAUAAAUGAGGU-AGAUAUUUGUUU----AACUG ------------........((((((-(........))(((...((((((((((((((((...))---))))))))))))))....)))-.......)))))----..... ( -28.70, z-score = -2.72, R) >droEre2.scaffold_4770 2933729 90 + 17746568 ------------GCGAAUUAAGCCAG-CCCAAGGACGCGCCGGGUUAUGCAAGUCUAUGCAACGA---AUGGGCUUGCAUAAUAGAGGU-AAAUAUUUGUUU----AACUG ------------((((((...(((.(-(((..((.....))))))(((((((((((((.......---))))))))))))).....)))-....))))))..----..... ( -28.90, z-score = -2.29, R) >droAna3.scaffold_13340 18326407 90 - 23697760 ------------GCGAAUUAAGCCGG-UCCAAGGACGCGCCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAAAGAC-AAAUAUUUGUUU----AACGG ------------((.......))(((-((....))).)).((((((((((((((((((.(...).---)))))))))))))))..((((-((....))))))----..))) ( -29.90, z-score = -2.68, R) >dp4.chr2 11107964 92 - 30794189 ------------GCGAAUUAAGCCAGUCCCAAGGCCGCGCCCGGUUAUGCAAGUCUAUGCAACGGG--AUGGGCUUGCAUAAUAAAGGC-AAAUAUUUGUUU----AACUG ------------(((......(((........))))))(((..(((((((((((((((.(....).--)))))))))))))))...)))-............----..... ( -32.80, z-score = -2.47, R) >droPer1.super_3 4946303 92 - 7375914 ------------GCGAAUUAAGCCAGUCCCAAGGCCGCGCCCGGUUAUGCAAGUCUAUGCAACGGG--AUGGGCUUGCAUAAUAAAGGC-AAAUAUUUGUUU----AACUG ------------(((......(((........))))))(((..(((((((((((((((.(....).--)))))))))))))))...)))-............----..... ( -32.80, z-score = -2.47, R) >droWil1.scaffold_181089 285832 109 - 12369635 --UCGAGUCCAAGGCCACCAACGGACCAACUUCGAGGCAUCCGGUUAUGCAAGUCUAUGCAAUUGCAAAUGGGUUUGCAUAAUAAAUGCCAAAUAUUUAUUUGUUUAACUU --(((((.....((((......)).))...)))))(((((...(((((((((..((((((....)))..)))..)))))))))..)))))(((((......)))))..... ( -32.30, z-score = -2.93, R) >droVir3.scaffold_13047 6025741 99 + 19223366 GCGAAUCUGUGAGUAGUCCGA--C----UCAAGGACGUGCCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAAAUGC-AAAUAUUGGUUUUA--AGCCG (((..(((.(((((......)--)----))).)))..)))...(((((((((((((((.(...).---)))))))))))))))......-.......(((....--.))). ( -33.00, z-score = -2.59, R) >droMoj3.scaffold_6540 16425762 90 + 34148556 ------------GCGAAUCUG--CAG-UUCAAGGACGUGCCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAGAUGC-AAAUAUUGGCUUUA--AGCCG ------------((..(((((--(((-((....))).)))...(((((((((((((((.(...).---)))))))))))))))))))))-.......(((....--.))). ( -31.30, z-score = -2.30, R) >droGri2.scaffold_14906 11373184 85 + 14172833 ------------GCGAAUC----C----UCAAGGACGUGGCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAAAUGC-GUAUAUUAGUUUUA--AGCCG ------------(((..((----(----....))))))(((..(((((((((((((((.(...).---)))))))))))))))....((-........))....--.))). ( -26.70, z-score = -1.89, R) >consensus ____________GCGAAUUAAGCCAG_CCCAAGGACGCGCCCGGUUAUGCAAGUCUAUGCAACGG___AUGGGCUUGCAUAAUAAAGGC_AAAUAUUUGUUU____AACUG ...........................................(((((((((((((((..........)))))))))))))))............................ (-15.24 = -15.04 + -0.20)

| Location | 2,683,509 – 2,683,600 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.92 |
| Shannon entropy | 0.42466 |
| G+C content | 0.43041 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
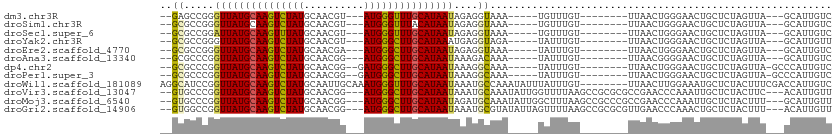
>dm3.chr3R 2683509 91 + 27905053 --GAGCCGGGUUAUGCAAGUCUAUGCAACGU---AUGGGUUUGCAUAAUAGAGGUAAA-----UGUUUGU--------UUAACUGGGAACUGCUCUAGUUA---GCAUUGUC --..(((..(((((((((..((((((...))---))))..)))))))))...))).((-----(((....--------.((((((((......))))))))---)))))... ( -31.10, z-score = -3.69, R) >droSim1.chr3R 2718247 91 + 27517382 --GCGCCGGGUUAUGCAAGUCUAUGCAACGU---AUGGGUUUACAUAAUAGAGGUAAA-----UGUUUGU--------UUAACUGGGAACUGCUCUAGUUA---GCAUUGUC --(((((..((((((.((..((((((...))---))))..)).))))))...)))...-----.......--------.((((((((......))))))))---))...... ( -25.30, z-score = -1.64, R) >droSec1.super_6 2774952 91 + 4358794 --GCGCCGGAUUAUGCAAGUUUAUGCAACGU---AUGGGUUUGCAUAAUAGAGGUAAA-----UGUUUGU--------UUAACUGGGAACUGCUCUAGUUA---GCAUUGUC --(((((..((((((((((..(((((...))---)))..))))))))))...)))...-----.......--------.((((((((......))))))))---))...... ( -29.10, z-score = -3.03, R) >droYak2.chr3R 21102845 91 - 28832112 --GCGCCGGGUUAUGCAAGUCUAUGCAACGU---AUGGGCUUGCAUAAAUGAGGUAGA-----UAUUUGU--------UUAACUGGGAACUGCUCUAGUUA---GCAUUGUU --(((((...((((((((((((((((...))---))))))))))))))....)))...-----.......--------.((((((((......))))))))---))...... ( -34.10, z-score = -3.96, R) >droEre2.scaffold_4770 2933752 91 + 17746568 --GCGCCGGGUUAUGCAAGUCUAUGCAACGA---AUGGGCUUGCAUAAUAGAGGUAAA-----UAUUUGU--------UUAACUGGGAACUGCUCUAGUUA---GCAUUGUC --(((((..(((((((((((((((.......---)))))))))))))))...)))...-----.......--------.((((((((......))))))))---))...... ( -33.60, z-score = -4.24, R) >droAna3.scaffold_13340 18326430 91 - 23697760 --GCGCCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAAAGACAAA-----UAUUUGU--------UUAACGGGGAACUGCUCUAGUUA---GCAUUGUC --((.(((((((((((((((((((.(...).---)))))))))))))))..((((((.-----...))))--------))..)))).(((((...))))).---))...... ( -32.40, z-score = -3.79, R) >dp4.chr2 11107988 94 - 30794189 --GCGCCCGGUUAUGCAAGUCUAUGCAACGG--GAUGGGCUUGCAUAAUAAAGGCAAA-----UAUUUGU--------UUAACUGGGAACUGCUCUAGUUA-GCCCAUUGUC --(((((..(((((((((((((((.(....)--.)))))))))))))))...)))...-----.......--------.((((((((......))))))))-))........ ( -35.30, z-score = -3.42, R) >droPer1.super_3 4946327 94 - 7375914 --GCGCCCGGUUAUGCAAGUCUAUGCAACGG--GAUGGGCUUGCAUAAUAAAGGCAAA-----UAUUUGU--------UUAACUGGGAACUGCUCUAGUUA-GCCCAUUGUC --(((((..(((((((((((((((.(....)--.)))))))))))))))...)))...-----.......--------.((((((((......))))))))-))........ ( -35.30, z-score = -3.42, R) >droWil1.scaffold_181089 285864 104 - 12369635 AGGCAUCCGGUUAUGCAAGUCUAUGCAAUUGCAAAUGGGUUUGCAUAAUAAAUGCCAAAUAUUUAUUUGU--------UUAACUUGGAAAUGCUCUACUUUCGACCAUUGUC .(((((...(((((((((..((((((....)))..)))..)))))))))..)))))(((((......)))--------))........((((.((.......)).))))... ( -27.30, z-score = -2.38, R) >droVir3.scaffold_13047 6025771 104 + 19223366 --GUGCCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAAAUGCAAAUAUUGGUUUUAAGCCGCGCGCCGAACCCAAAUUGCUCUACUUC---ACAUUGUU --(((....(((((((((((((((.(...).---)))))))))))))))....((((.....(((((...((...))...)))))....)))).......)---))...... ( -29.10, z-score = -0.82, R) >droMoj3.scaffold_6540 16425783 104 + 34148556 --GUGCCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAGAUGCAAAUAUUGGCUUUAAGCCGCCCGCCGAACCCAAAUUGCUCUACUUU---GCAUUGUU --......((((.((((......)))).(((---.(((((((((((.....)))))).....(((.....))))))))))))))).....(((........---)))..... ( -32.60, z-score = -1.49, R) >droGri2.scaffold_14906 11373200 104 + 14172833 --GUGGCCGGUUAUGCAAGUCUAUGCAACGG---AUGGGCUUGCAUAAUAAAUGCGUAUAUUAGUUUUAAGCCGCGCGUUGAACCCAAACUGCUCUACUUU---ACAUUGUU --(((((..(((((((((((((((.(...).---)))))))))))))))....((........)).....)))))(((((.......))).))........---........ ( -29.10, z-score = -0.89, R) >consensus __GCGCCCGGUUAUGCAAGUCUAUGCAACGG___AUGGGCUUGCAUAAUAAAGGCAAA_____UAUUUGU________UUAACUGGGAACUGCUCUAGUUA___GCAUUGUC ....((...(((((((((((((((..........)))))))))))))))....))......................................................... (-15.52 = -15.30 + -0.22)
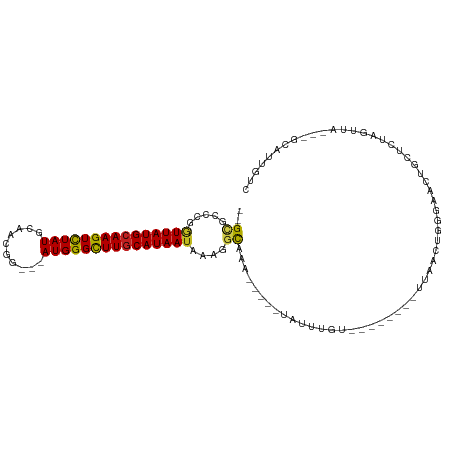
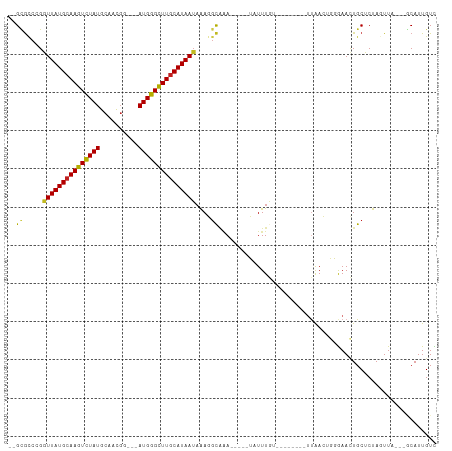
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:46 2011