| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,674,531 – 2,674,646 |
| Length | 115 |
| Max. P | 0.944920 |

| Location | 2,674,531 – 2,674,646 |
|---|---|
| Length | 115 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 61.35 |
| Shannon entropy | 0.80828 |
| G+C content | 0.46660 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.60 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.944920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
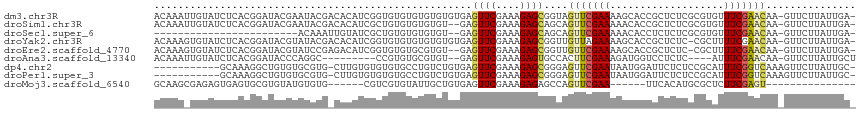
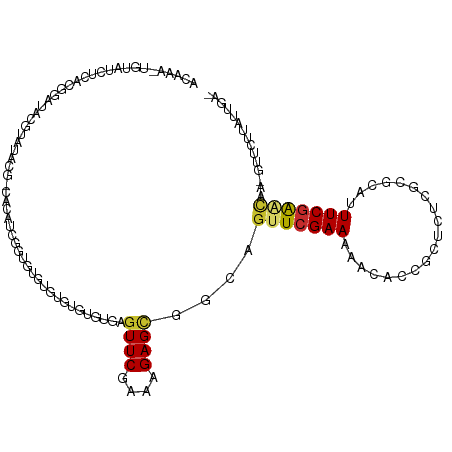
>dm3.chr3R 2674531 115 + 27905053 ACAAAUUGUAUCUCACGGAUACGAAUACGACACAUCGGUGUGUGUGUGUGUGAGUUCGAAAGAGCGGUAGUUCGAAAAGCACCGCUCUCGCGUGUUUCGAACAA-GUUCUUAUUGA- .(((.(((...((((((.(((((.((((((....)).)))).))))).))))))..)))((((((....(((((((..((((.((....)))))))))))))..-)))))).))).- ( -37.10, z-score = -1.68, R) >droSim1.chr3R 2709145 113 + 27517382 ACAAAUUGUAUCUCACGGAUACGAAUACGACACAUCGCUGUGUGUGUGU--GAGUUCGAAAGAGCAGCAGUUCGAAAAACACCGCUCUCGCGUGUUUCGAACAA-GUUCUUAUUGA- .....(((((((.....)))))))((((.((((((....))))))))))--..((((....))))....(((((((..((((.((....)))))))))))))..-...........- ( -34.90, z-score = -1.62, R) >droSec1.super_6 2765913 89 + 4358794 ------------------------ACAAAUUGUAUCGCUGUGUGUGUGU--GAGUUCGAAAGAGCAGCAGUUCGAAAAACACCUCUCUCGCGUGUUUCGAACAA-GUUCUUAUUGA- ------------------------.(((..(((.((((.........))--))((((....)))).)))(((((((..(((((......).)))))))))))..-.......))).- ( -22.40, z-score = -0.73, R) >droYak2.chr3R 21093690 114 - 28832112 ACAAAGUGUAUCUCACGGAUACGUAUACGACACAUCGGUGUGUGUGUGUGUGAGUUCGAAAGAGCGGUUGUUAGAAAAGCACCGCUCUC-CGCUUUUCGAACAA-GUUCUUAUUGA- .(((........(((((.((((((((((((....)).)))))))))).)))))(((((((((((((((.(((.....))))))))))).-.....)))))))..-.......))).- ( -37.20, z-score = -2.24, R) >droEre2.scaffold_4770 2924068 112 + 17746568 ACAAAGUGUAUCUCACGGAUACGUAUCCGAGACAUCGGUGUGUGCGUGU--GAGUUCGAAAGAGCGGUUGUUCGAAAAGCACCGCUCUC-CGCUUUUCGAACAA-GUUCUUAUUGA- .(((......((.((((.(((((...((((....))))))))).)))).--))((((....))))..(((((((((((((.........-.)))))))))))))-.......))).- ( -41.10, z-score = -3.30, R) >droAna3.scaffold_13340 18318302 101 - 23697760 ACAAAUUGUAUCUCACGGAUACCCAGGC---------CCGUGUGCGUGU--GAGUUCGAAAGAGUGCCACUUCGAAAGAUGGUCCUCUC----AUUUCGAACAA-GUUCUUAUUGCU .......(((((.((((.((((......---------..)))).)))).--))(((((((((((..(((..((....)))))..)))).----..)))))))..-........))). ( -32.30, z-score = -3.02, R) >dp4.chr2 11099066 104 - 30794189 -----------GCAAAGGCUGUGUGCGUG-CUUGUGUGUGUGCCUGUCUGUGAGUUCGAAAGAGCGGGAGUUCGAAUAAUGGAUUCUCUCCGCAUUUCGGUCAAAGUUCUUAUUGC- -----------((((((((.(.(..((..-(....)..))..)).))))(((.((((....))))(((((((((.....)))))))))..)))...................))))- ( -31.60, z-score = -1.68, R) >droPer1.super_3 4937356 104 - 7375914 -----------GCAAAGGCUGUGUGCGUG-CUUGUGUGUGUGCCUGUCUGUGAGUUCGAAAGAGCGGGAGUUCGAAUAAUGGAUUCUCUCCGCAUUUCGGUCAAAGUUCUUAUUGC- -----------((((((((.(.(..((..-(....)..))..)).))))(((.((((....))))(((((((((.....)))))))))..)))...................))))- ( -31.60, z-score = -1.68, R) >droMoj3.scaffold_6540 17697336 90 - 34148556 GCAAGCGAGAGUGAGUGCGUGUAUGUGUG------CGUCGUGUAUUGCUGUGAGUUCGAAAGAGAGCCAGUUCGAA------UUCACAUGCGCUCUUCGAGU--------------- (((.((....))...)))(((((((((..------..(((......((((.(..(((....)))..))))).))).------..))))))))).........--------------- ( -31.30, z-score = -1.78, R) >consensus ACAAA_UGUAUCUCACGGAUACGUAUACG_CACAUCGGUGUGUGUGUGUGUGAGUUCGAAAGAGCGGCAGUUCGAAAAACACCGCUCUCGCGCAUUUCGAACAA_GUUCUUAUUGA_ .....................................................((((....))))....(((((((...................)))))))............... ( -9.46 = -9.60 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:44 2011