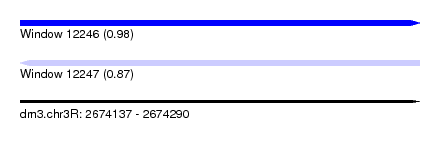
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,674,137 – 2,674,290 |
| Length | 153 |
| Max. P | 0.976843 |

| Location | 2,674,137 – 2,674,290 |
|---|---|
| Length | 153 |
| Sequences | 8 |
| Columns | 177 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.47862 |
| G+C content | 0.42284 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -24.29 |
| Energy contribution | -26.54 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
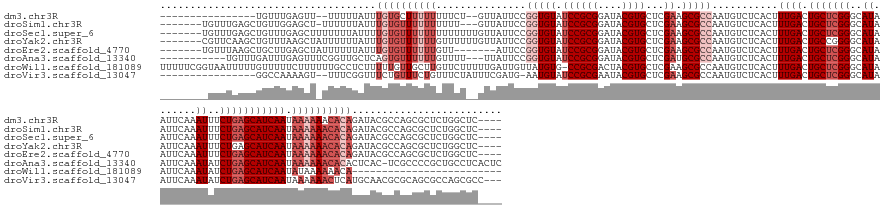
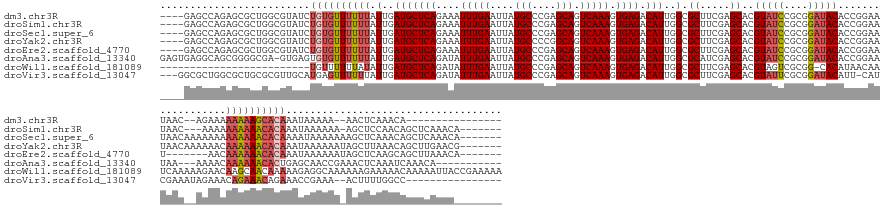
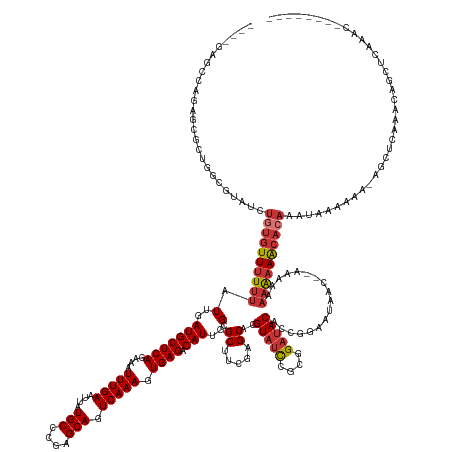
>dm3.chr3R 2674137 153 + 27905053 ----------------UGUUUGAGUU--UUUUUAUUUGUGCUUUUUUUUCU--GUUAUUCCGGUGUAUCCGCGGAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUUUCUGAGCAUCAAUAAAAAACACAGAUACGCCAGCGCUCUGGCUC---- ----------------((((((.(((--((((.(((.((((((........--..((((((((.....))).))))).((((((((.(((.....(((.......)))))))))))))).............)))))).)))))))))).)))))).(((((....)))))..---- ( -44.90, z-score = -2.76, R) >droSim1.chr3R 2708742 162 + 27517382 -------UGUUUGAGCUGUUGGAGCU-UUUUUUAUUUGUGUUUUUUUUUU---GUUAUUCCGGUGUAUCCGCGGAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUUUCUGAGCAUCAAUAAAAAACACAGAUACGCCAGCGCUCUGGCUC---- -------.((..((((.(((((.(..-.....((((((((((((((....---........(((((.((((((....))))...)).)))))...........((((.((((((((.((.......)).)))))))))))).))))))))))))))).)))))))))..))..---- ( -50.41, z-score = -2.90, R) >droSec1.super_6 2765506 166 + 4358794 -------UGUUUGAGCUGUUUGAGCUUUUUUUAUUUUGUGUUUUUUUUUUUUUGUUAUUCCGGUGUAUCCGCGGAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUUUCUGAGCAUCAAUAAAAAACACAGAUACGCCAGCGCUCUGGCUC---- -------.....(((((....((((.........((((((((((((...............(((((.((((((....))))...)).)))))...........((((.((((((((.((.......)).)))))))))))).)))))))))))).........)))).)))))---- ( -45.97, z-score = -2.03, R) >droYak2.chr3R 21093299 166 - 28832112 -------CGUUCAAGCUGUUUAAGCUAUUUUUUAUUUGUGUUUUUUGUUUUUUGUUAUUCCGGUGUAUCCGCGGAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCCGGGGCAUAAUUCAAAUUUCUGAGCAUCAAUAAAAAACACAGAUACGCCAGCGCUCUGGCUC---- -------......((((.....))))......(((((((((((((((((...(((..((((((((((((....)))))((((.....))))....(((.......))).)))))))......(((......))))))..))))))))))))))))).(((((....)))))..---- ( -50.70, z-score = -3.31, R) >droEre2.scaffold_4770 2923668 159 + 17746568 -------UGUUUAAGCUGCUUGAGCUAUUUUUUAUUUGUGUUUUUUGUU-------AUUCCGGUGUAUCCGCGGAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUUUCUGAGCAUCAAUAAAAAACACAGAUACGCCAGCGCUCUGGCUC---- -------.(((((((...))))))).......(((((((((((((((((-------.....(((((.((((((....))))...)).)))))...(((.......)))((((((((.((.......)).))))))))..))))))))))))))))).(((((....)))))..---- ( -51.00, z-score = -3.50, R) >droAna3.scaffold_13340 18317807 162 - 23697760 -----------UGUUUGAUUUGAGUUUCGGUUGCUCAGUGUUUUUUGUUUU---UUAUUCCGGUGUAUCCGCGGAUACGUGCUCGAUGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUAUCUGAGCAUCAAUAAAAAACACACUCAC-UCGCCCCGCUGCCUCACUC -----------.........(((((..(((..((..((((.....((((((---(((((..((((((((((((....))))...))))))))...(((.......)))(((((((..((.......))..)))))))..)))))))))))....)))-).)).)))..).))))... ( -47.10, z-score = -3.03, R) >droWil1.scaffold_181089 12059214 151 + 12369635 UUUUUCGGUAAUUUUUGUUUUUCUUUUUUGCCUCUUUUUGUUGCUUGUUCUUUUUGAUUGUUAUGUG-CCGCGACUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUAUCUGAGCAUCAAUAUAAAAAACA------------------------- ......(((((................)))))..(((.(((((..(((((..(((((......((((-((.(((....((((.....))))....(((.......)))...)))))))))..))))).....))))).))))).))).....------------------------- ( -30.59, z-score = -0.92, R) >droVir3.scaffold_13047 13173293 155 - 19223366 ----------------GGCCAAAAGU--UUUCGGUUUCUGUUUCUGUUUCUAUUUCGAUG-AAUGUAUCCGCGAAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUAUCUGAGCAUCAAUAAAAAACUCAUGCAACGCGCAGCGCCAGCGCC--- ----------------((((....(.--...)(((..((((............(((((..-.((((((......))))))..)))))(((...(((.......((((.(((((((..((.......))..))))))))))).........)))....))))))).))).).)))--- ( -34.09, z-score = 1.07, R) >consensus ________GUUUGAGCUGUUUGAGCU_UUUUUUAUUUGUGUUUUUUGUUUU__GUUAUUCCGGUGUAUCCGCGGAUACGUGCUCGAAGCGCCAAUGUCUCACUUUGACUGCUCGGGCAUAAUUCAAAUUUCUGAGCAUCAAUAAAAAACACAGAUACGCCAGCGCUCUGGCUC____ ....................................((((((((((...............(((((.((((((....))))...)).)))))...........((((.(((((((..((.......))..))))))))))).))))))))))......................... (-24.29 = -26.54 + 2.25)
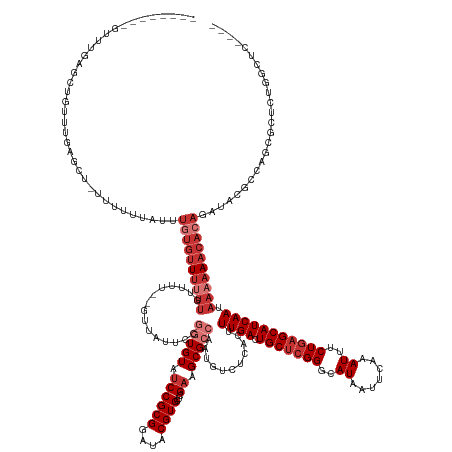
| Location | 2,674,137 – 2,674,290 |
|---|---|
| Length | 153 |
| Sequences | 8 |
| Columns | 177 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.47862 |
| G+C content | 0.42284 |
| Mean single sequence MFE | -40.59 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.76 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2674137 153 - 27905053 ----GAGCCAGAGCGCUGGCGUAUCUGUGUUUUUUAUUGAUGCUCAGAAAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUCCGCGGAUACACCGGAAUAAC--AGAAAAAAAAGCACAAAUAAAAA--AACUCAAACA---------------- ----..(((((....))))).(((.((((((((((...((((((((....(((((....(((....))).))))).)))).))))((.((.....))..(((((....))))).))........--.....)))))))))).)))....--..........---------------- ( -40.00, z-score = -1.62, R) >droSim1.chr3R 2708742 162 - 27517382 ----GAGCCAGAGCGCUGGCGUAUCUGUGUUUUUUAUUGAUGCUCAGAAAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUCCGCGGAUACACCGGAAUAAC---AAAAAAAAAACACAAAUAAAAAA-AGCUCCAACAGCUCAAACA------- ----((((..((((.......(((.((((((((((...((((((((....(((((....(((....))).))))).)))).))))((.((.....))..(((((....))))).))........---....)))))))))).))).....-.)))).....)))).....------- ( -43.02, z-score = -1.96, R) >droSec1.super_6 2765506 166 - 4358794 ----GAGCCAGAGCGCUGGCGUAUCUGUGUUUUUUAUUGAUGCUCAGAAAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUCCGCGGAUACACCGGAAUAACAAAAAAAAAAAAACACAAAAUAAAAAAAGCUCAAACAGCUCAAACA------- ----((((..((((((((((((....(((((((...(((((((((.(..((.......))..).))))).))))...)))))))..)))))...)))..(((((....))))).......................................)))).....)))).....------- ( -42.20, z-score = -1.91, R) >droYak2.chr3R 21093299 166 + 28832112 ----GAGCCAGAGCGCUGGCGUAUCUGUGUUUUUUAUUGAUGCUCAGAAAUUUGAAUUAUGCCCCGGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUCCGCGGAUACACCGGAAUAACAAAAAACAAAAAACACAAAUAAAAAAUAGCUUAAACAGCUUGAACG------- ----..(((((....))))).(((.((((((((((...((((((((....(((((....(((....))).))))).)))).))))((.((.....))..(((((....))))).))...............)))))))))).)))......((((.....))))......------- ( -43.80, z-score = -2.02, R) >droEre2.scaffold_4770 2923668 159 - 17746568 ----GAGCCAGAGCGCUGGCGUAUCUGUGUUUUUUAUUGAUGCUCAGAAAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUCCGCGGAUACACCGGAAU-------AACAAAAAACACAAAUAAAAAAUAGCUCAAGCAGCUUAAACA------- ----..(((((....))))).(((.((((((((((...((((((((....(((((....(((....))).))))).)))).))))((.((.....))..(((((....))))).)).....-------...)))))))))).)))......((((.....))))......------- ( -41.90, z-score = -1.30, R) >droAna3.scaffold_13340 18317807 162 + 23697760 GAGUGAGGCAGCGGGGCGA-GUGAGUGUGUUUUUUAUUGAUGCUCAGAUAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCAUCGAGCACGUAUCCGCGGAUACACCGGAAUAA---AAAACAAAAAACACUGAGCAACCGAAACUCAAAUCAAACA----------- ((.((((....(((.((.(-(((....(((((((((((..(((((.(((......)))((((((((...(((.......))).)))).)))).))))).(((((....))))).....)))))---)))))).....))))..))..)))...))))..)).....----------- ( -46.60, z-score = -1.98, R) >droWil1.scaffold_181089 12059214 151 - 12369635 -------------------------UGUUUUUUAUAUUGAUGCUCAGAUAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAGUCGCGG-CACAUAACAAUCAAAAAGAACAAGCAACAAAAAGAGGCAAAAAAGAAAAACAAAAAUUACCGAAAAA -------------------------((((((((...(..(((((((....(((((....(((....))).))))).)))).)))..).(((((..((.(((....))))-).........((.....))..............)))))......))))))))............... ( -30.20, z-score = -1.15, R) >droVir3.scaffold_13047 13173293 155 + 19223366 ---GGCGCUGGCGCUGCGCGUUGCAUGAGUUUUUUAUUGAUGCUCAGAUAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUUCGCGGAUACAUU-CAUCGAAAUAGAAACAGAAACAGAAACCGAAA--ACUUUUGGCC---------------- ---(((.((((((((((.....))....(((((...(((((((((.(.(((.......))).).))))).))))...)))))...)))))(((((....(((((....)))))...-..))))).......)))...(((((.......--..))))))))---------------- ( -37.00, z-score = 1.29, R) >consensus ____GAGCCAGAGCGCUGGCGUAUCUGUGUUUUUUAUUGAUGCUCAGAAAUUUGAAUUAUGCCCGAGCAGUCAAAGUGAGACAUUGGCGCUUCGAGCACGUAUCCGCGGAUACACCGGAAUAAC__AAAAAAAAAAACACAAAUAAAAAA_AGCUCAAACAGCUCAAAC________ .........................((((((((((.(..(((((((....(((((....(((....))).))))).)))).)))..).((.....))..(((((....)))))..................)))))))))).................................... (-26.00 = -26.76 + 0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:43 2011