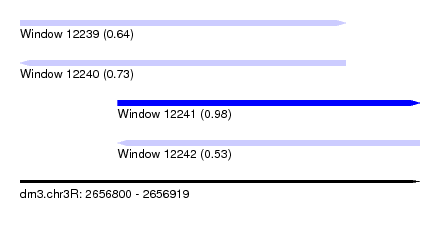
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,656,800 – 2,656,919 |
| Length | 119 |
| Max. P | 0.984313 |

| Location | 2,656,800 – 2,656,897 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 127 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Shannon entropy | 0.37467 |
| G+C content | 0.32013 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -13.94 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
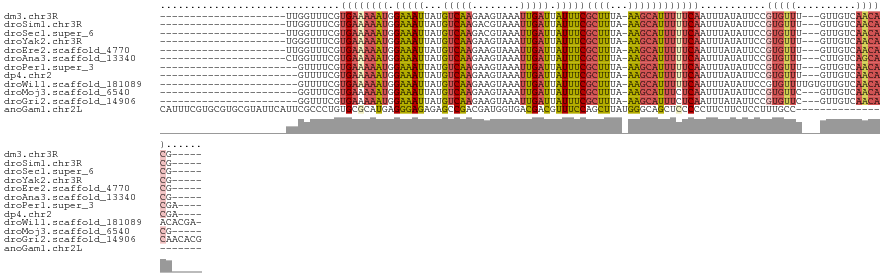
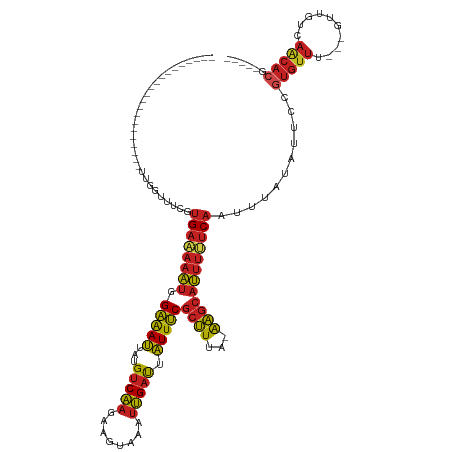
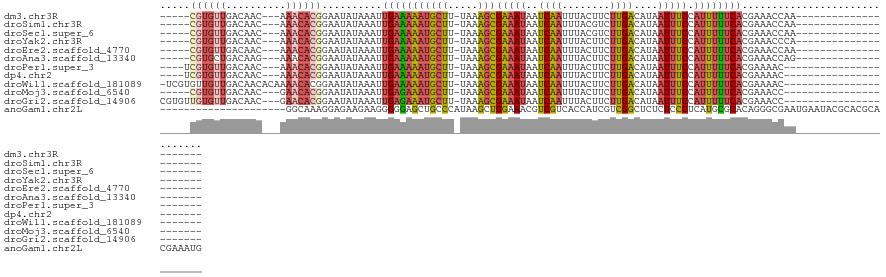
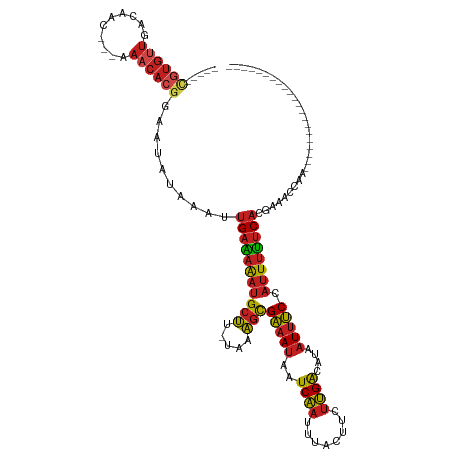
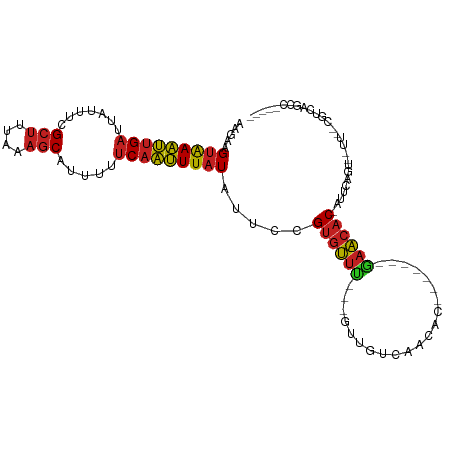
>dm3.chr3R 2656800 97 + 27905053 ---------------------UUGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG----- ---------------------..((.....((((((((.(((((...(((((........))))).)))))(((...-.))))))))))).........))(((((.---......))))).----- ( -18.74, z-score = -1.34, R) >droSim1.chr3R 2691506 97 + 27517382 ---------------------UUGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGACGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG----- ---------------------..((.....((((((((.(((((...(((((........))))).)))))(((...-.))))))))))).........))(((((.---......))))).----- ( -18.74, z-score = -1.04, R) >droSec1.super_6 2748457 97 + 4358794 ---------------------UUGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGACGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG----- ---------------------..((.....((((((((.(((((...(((((........))))).)))))(((...-.))))))))))).........))(((((.---......))))).----- ( -18.74, z-score = -1.04, R) >droYak2.chr3R 21075673 97 - 28832112 ---------------------UGGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG----- ---------------------.(((.....((((((((.(((((...(((((........))))).)))))(((...-.)))))))))))........)))(((((.---......))))).----- ( -20.12, z-score = -1.70, R) >droEre2.scaffold_4770 2906074 97 + 17746568 ---------------------UUGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG----- ---------------------..((.....((((((((.(((((...(((((........))))).)))))(((...-.))))))))))).........))(((((.---......))))).----- ( -18.74, z-score = -1.34, R) >droAna3.scaffold_13340 18301399 97 - 23697760 ---------------------CUGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---CUUGUCAGCACG----- ---------------------..((.....((((((((.(((((...(((((........))))).)))))(((...-.))))))))))).........))(((((.---......))))).----- ( -18.74, z-score = -1.48, R) >droPer1.super_3 4919143 96 - 7375914 -----------------------GUUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACGA---- -----------------------.......((((((((.(((((...(((((........))))).)))))(((...-.)))))))))))..........((((((.---......)))))).---- ( -19.80, z-score = -1.84, R) >dp4.chr2 11080799 96 - 30794189 -----------------------GUUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACGA---- -----------------------.......((((((((.(((((...(((((........))))).)))))(((...-.)))))))))))..........((((((.---......)))))).---- ( -19.80, z-score = -1.84, R) >droWil1.scaffold_181089 12036476 102 + 12369635 -----------------------GUUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUUUCAAUUUAUAUUCCGUGUUUUGUGUUGUCAACAACACGA- -----------------------.......((((((((.(((((...(((((........))))).)))))(((...-.)))))))))))................((((((((....))))))))- ( -22.60, z-score = -2.20, R) >droMoj3.scaffold_6540 17673531 95 - 34148556 -----------------------GGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUCUCAAUUUAUAUUCCGUGUUC---GUUGUCAACACG----- -----------------------((.(((.(((..(((....)))...))).)))(((((((((.......(((...-.))).....)))))))))...))(((((.---......))))).----- ( -15.90, z-score = -0.67, R) >droGri2.scaffold_14906 2756261 100 - 14172833 -----------------------GGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA-AAGCAUUUCUCAAUUUAUAUUCCGUGUUC---GUUGUCAACACAACACG -----------------------.....((((......((((..........(((((((.....)))))))(((...-.)))...............))))(((((.---......)))))..)))) ( -18.00, z-score = -1.03, R) >anoGam1.chr2L 3363059 106 + 48795086 CAUUUCGUGCGUGCGUAUUCAUUCGCCCUGUGCGCAUGAGGGAGAGAGCCGACGAUGGUGACGACGUUUCCAGCUUAUGGGCAGCUCCCCCUUCUUCUCCUUUGCC--------------------- ....(((((((..((.............))..)))))))((((((((((.((((.((....)).))))....))))..(((......))).....)))))).....--------------------- ( -33.52, z-score = -0.44, R) >consensus _____________________UUGGUUUCGUGAAAAAUGGAAAUUAUGUCAAGAAGUAAAUUGAUUAUUUCGCUUUA_AAGCAUUUUUCAAUUUAUAUUCCGUGUUU___GUUGUCAACACG_____ ..............................((((((((.(((((...(((((........))))).)))))((((...))))))))))))...........(((((..........)))))...... (-13.94 = -14.17 + 0.23)
| Location | 2,656,800 – 2,656,897 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Shannon entropy | 0.37467 |
| G+C content | 0.32013 |
| Mean single sequence MFE | -16.99 |
| Consensus MFE | -10.96 |
| Energy contribution | -11.45 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2656800 97 - 27905053 -----CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACCAA--------------------- -----((((((......---.))))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).........--------------------- ( -15.90, z-score = -2.16, R) >droSim1.chr3R 2691506 97 - 27517382 -----CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACGUCUUGACAUAAUUUCCAUUUUUCACGAAACCAA--------------------- -----((((((......---.))))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).........--------------------- ( -15.90, z-score = -1.56, R) >droSec1.super_6 2748457 97 - 4358794 -----CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACGUCUUGACAUAAUUUCCAUUUUUCACGAAACCAA--------------------- -----((((((......---.))))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).........--------------------- ( -15.90, z-score = -1.56, R) >droYak2.chr3R 21075673 97 + 28832112 -----CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACCCA--------------------- -----((((((......---.))))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).........--------------------- ( -15.90, z-score = -2.23, R) >droEre2.scaffold_4770 2906074 97 - 17746568 -----CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACCAA--------------------- -----((((((......---.))))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).........--------------------- ( -15.90, z-score = -2.16, R) >droAna3.scaffold_13340 18301399 97 + 23697760 -----CGUGCUGACAAG---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACCAG--------------------- -----((((((....))---...))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).........--------------------- ( -12.00, z-score = -0.67, R) >droPer1.super_3 4919143 96 + 7375914 ----UCGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAAAC----------------------- ----(((((((......---.))))))).........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).......----------------------- ( -16.40, z-score = -2.35, R) >dp4.chr2 11080799 96 + 30794189 ----UCGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAAAC----------------------- ----(((((((......---.))))))).........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).......----------------------- ( -16.40, z-score = -2.35, R) >droWil1.scaffold_181089 12036476 102 - 12369635 -UCGUGUUGUUGACAACACAAAACACGGAAUAUAAAUUGAAAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAAAC----------------------- -((((((((....))))))..................(((((((((((.-...)))(((((..((((........))))....))))).)))))))).))....----------------------- ( -17.20, z-score = -1.89, R) >droMoj3.scaffold_6540 17673531 95 + 34148556 -----CGUGUUGACAAC---GAACACGGAAUAUAAAUUGAGAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACC----------------------- -----((((((......---.))))))..........(((((((((((.-...)))(((((..((((........))))....))))).)))))))).......----------------------- ( -15.30, z-score = -1.46, R) >droGri2.scaffold_14906 2756261 100 + 14172833 CGUGUUGUGUUGACAAC---GAACACGGAAUAUAAAUUGAGAAAUGCUU-UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACC----------------------- ((((..(((((......---.)))))((((......(..((((..(((.-...)))((.....)).......))))..)......))))......)))).....----------------------- ( -18.10, z-score = -1.36, R) >anoGam1.chr2L 3363059 106 - 48795086 ---------------------GGCAAAGGAGAAGAAGGGGGAGCUGCCCAUAAGCUGGAAACGUCGUCACCAUCGUCGGCUCUCUCCCUCAUGCGCACAGGGCGAAUGAAUACGCACGCACGAAAUG ---------------------.((............(((((((..(((........(....)(.((.......)).)))).)))))))((((.(((.....))).))))....))............ ( -29.00, z-score = 0.81, R) >consensus _____CGUGUUGACAAC___AAACACGGAAUAUAAAUUGAAAAAUGCUU_UAAAGCGAAAUAAUCAAUUUACUUCUUGACAUAAUUUCCAUUUUUCACGAAACCAA_____________________ .....((((((..........))))))..........(((((((((((.....)))(((((..((((........))))....))))).)))))))).............................. (-10.96 = -11.45 + 0.49)
| Location | 2,656,829 – 2,656,919 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Shannon entropy | 0.43529 |
| G+C content | 0.35027 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.62 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
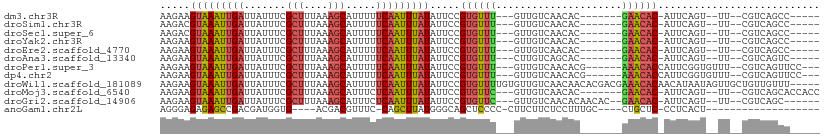
>dm3.chr3R 2656829 90 + 27905053 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACAC-------GAACAC-AUUCAGU--UU--CGUCAGCC----- ..((((((((((((.......(((....))).....))))))))).....((((((---((.(....)))-------))))))-.)))...--..--........----- ( -18.10, z-score = -2.92, R) >droSim1.chr3R 2691535 90 + 27517382 AAGACGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACAC-------GAACAC-AUUCAGU--UU--CGUCAGCC----- ..((((.(((((((.......(((....)))...................((((((---((.(....)))-------))))))-..)))))--))--))))....----- ( -22.60, z-score = -4.48, R) >droSec1.super_6 2748486 90 + 4358794 AAGACGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACAC-------GAACAC-AUUCAGU--UU--CGUCAGCC----- ..((((.(((((((.......(((....)))...................((((((---((.(....)))-------))))))-..)))))--))--))))....----- ( -22.60, z-score = -4.48, R) >droYak2.chr3R 21075702 90 - 28832112 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACAC-------GAACAC-AUUCAGU--UU--CGUCAGCC----- ..((((((((((((.......(((....))).....))))))))).....((((((---((.(....)))-------))))))-.)))...--..--........----- ( -18.10, z-score = -2.92, R) >droEre2.scaffold_4770 2906103 90 + 17746568 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACAC-------GAACAC-AUUCAGU--UU--CGUCAGCC----- ..((((((((((((.......(((....))).....))))))))).....((((((---((.(....)))-------))))))-.)))...--..--........----- ( -18.10, z-score = -2.92, R) >droAna3.scaffold_13340 18301428 90 - 23697760 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---CUUGUCAGCAC-------GAACAC-AUUCAGU--UU--CGUCAGUC----- ..((((((((((((.......(((....))).....))))))))).....((((((---..((....)).-------))))))-.)))...--..--........----- ( -14.90, z-score = -1.94, R) >droPer1.super_3 4919170 96 - 7375914 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG------AAACACCAUUCGGUGUUU--CGUCAGUUCC--- ..((((((((((((.......(((....))).....))))))))).))).(((((.---......)))))(------(((((((....)))))))--).........--- ( -24.10, z-score = -4.28, R) >dp4.chr2 11080826 96 - 30794189 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU---GUUGUCAACACG------AAACACCAUUCGGUGUUU--CGUCAGUUCC--- ..((((((((((((.......(((....))).....))))))))).))).(((((.---......)))))(------(((((((....)))))))--).........--- ( -24.10, z-score = -4.28, R) >droWil1.scaffold_181089 12036503 105 + 12369635 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUUUGUGUUGUCAACAACACGACGAAACACAACAUAAUAGUUGCUGUUGUUU----- ..((((((((((((.......(((....))).....))))))))).))).(((((((((((((....))))))...)))))))...(((((((...)))))))..----- ( -24.30, z-score = -2.43, R) >droMoj3.scaffold_6540 17673558 95 - 34148556 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUCUCAAUUUAUAUUCCGUGUUC---GUUGUCAACAC-------GAACAC-AUUCAGU--UU--CGUCAGCACCACC ..((((((((((((.......(((....))).....))))))))).....((((((---((.(....)))-------))))))-.)))...--..--............. ( -19.50, z-score = -3.57, R) >droGri2.scaffold_14906 2756288 94 - 14172833 AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUCUCAAUUUAUAUUCCGUGUUC---GUUGUCAACACAACAC--GAACAC-AUUCAGU--UU--CGUCAGC------ ..((((((((((((.......(((....))).....))))))))).....((((((---(((((....)))..))--))))))-.)))...--..--.......------ ( -19.90, z-score = -3.44, R) >anoGam1.chr2L 3363097 80 + 48795086 AGGGAGAGAGCCGACGAUGGUG----ACGACGUUUC-CAGCUUAUGGGCAGCUCCCC-CUUCUUCUCCUUUGC----CUGCUC-CCUCACU------------------- ((((((.((((.((((.((...----.)).))))..-..))))..((((((......-...........))))----)).)))-)))....------------------- ( -23.83, z-score = -0.05, R) >consensus AAGAAGUAAAUUGAUUAUUUCGCUUUAAAGCAUUUUUCAAUUUAUAUUCCGUGUUU___GUUGUCAACAC_______GAACAC_AUUCAGU__UU__CGUCAGCC_____ .....(((((((((.......(((....))).....))))))))).....((((((.....((....))........))))))........................... ( -8.53 = -8.62 + 0.09)
| Location | 2,656,829 – 2,656,919 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Shannon entropy | 0.43529 |
| G+C content | 0.35027 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -7.76 |
| Energy contribution | -8.63 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
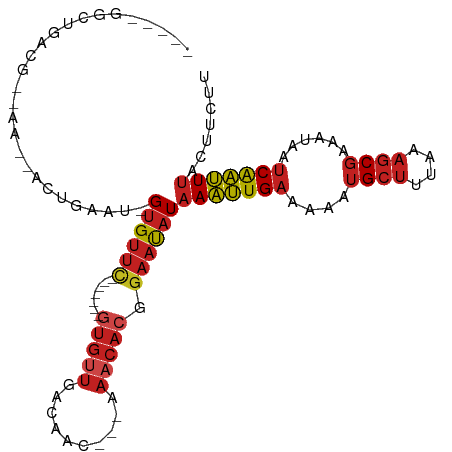
>dm3.chr3R 2656829 90 - 27905053 -----GGCUGACG--AA--ACUGAAU-GUGUUC-------GUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU -----.......(--((--.......-...(((-------(((((......---.))))))))...((((((((....((((....))))......)))))))).))).. ( -15.80, z-score = -1.52, R) >droSim1.chr3R 2691535 90 - 27517382 -----GGCUGACG--AA--ACUGAAU-GUGUUC-------GUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACGUCUU -----....((((--..--.......-...(((-------(((((......---.))))))))...((((((((....((((....))))......)))))))))))).. ( -20.40, z-score = -2.77, R) >droSec1.super_6 2748486 90 - 4358794 -----GGCUGACG--AA--ACUGAAU-GUGUUC-------GUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACGUCUU -----....((((--..--.......-...(((-------(((((......---.))))))))...((((((((....((((....))))......)))))))))))).. ( -20.40, z-score = -2.77, R) >droYak2.chr3R 21075702 90 + 28832112 -----GGCUGACG--AA--ACUGAAU-GUGUUC-------GUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU -----.......(--((--.......-...(((-------(((((......---.))))))))...((((((((....((((....))))......)))))))).))).. ( -15.80, z-score = -1.52, R) >droEre2.scaffold_4770 2906103 90 - 17746568 -----GGCUGACG--AA--ACUGAAU-GUGUUC-------GUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU -----.......(--((--.......-...(((-------(((((......---.))))))))...((((((((....((((....))))......)))))))).))).. ( -15.80, z-score = -1.52, R) >droAna3.scaffold_13340 18301428 90 + 23697760 -----GACUGACG--AA--ACUGAAU-GUGUUC-------GUGCUGACAAG---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU -----..((((((--((--((.....-)).)))-------)).((....))---.....)))....((((((((....((((....))))......))))))))...... ( -12.10, z-score = -0.53, R) >droPer1.super_3 4919170 96 + 7375914 ---GGAACUGACG--AAACACCGAAUGGUGUUU------CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU ---((((((((((--(((((((....)))))))------)))(((......---.))).)))....((((((((....((((....))))......)))))))).)))). ( -24.20, z-score = -4.04, R) >dp4.chr2 11080826 96 + 30794189 ---GGAACUGACG--AAACACCGAAUGGUGUUU------CGUGUUGACAAC---AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU ---((((((((((--(((((((....)))))))------)))(((......---.))).)))....((((((((....((((....))))......)))))))).)))). ( -24.20, z-score = -4.04, R) >droWil1.scaffold_181089 12036503 105 - 12369635 -----AAACAACAGCAACUAUUAUGUUGUGUUUCGUCGUGUUGUUGACAACACAAAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU -----........(((((......)))))(((((((.((((((....)))))).....))))))).((((((((....((((....))))......))))))))...... ( -22.60, z-score = -2.33, R) >droMoj3.scaffold_6540 17673558 95 + 34148556 GGUGGUGCUGACG--AA--ACUGAAU-GUGUUC-------GUGUUGACAAC---GAACACGGAAUAUAAAUUGAGAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU (((..((....))--..--)))....-((((((-------((.......))---))))))((((..((((((((....((((....))))......)))))))).)))). ( -21.20, z-score = -2.02, R) >droGri2.scaffold_14906 2756288 94 + 14172833 ------GCUGACG--AA--ACUGAAU-GUGUUC--GUGUUGUGUUGACAAC---GAACACGGAAUAUAAAUUGAGAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU ------.......--..--.......-((((((--(((((.....))).))---))))))((((..((((((((....((((....))))......)))))))).)))). ( -21.70, z-score = -2.44, R) >anoGam1.chr2L 3363097 80 - 48795086 -------------------AGUGAGG-GAGCAG----GCAAAGGAGAAGAAG-GGGGAGCUGCCCAUAAGCUG-GAAACGUCGU----CACCAUCGUCGGCUCUCUCCCU -------------------((.((((-((((.(----((...((.((.((..-(((......))).......(-....).)).)----).))...))).)))))))).)) ( -27.00, z-score = -0.11, R) >consensus _____GGCUGACG__AA__ACUGAAU_GUGUUC_______GUGUUGACAAC___AAACACGGAAUAUAAAUUGAAAAAUGCUUUAAAGCGAAAUAAUCAAUUUACUUCUU ........................................(((((..........)))))(((...((((((((....((((....))))......))))))))..))). ( -7.76 = -8.63 + 0.87)
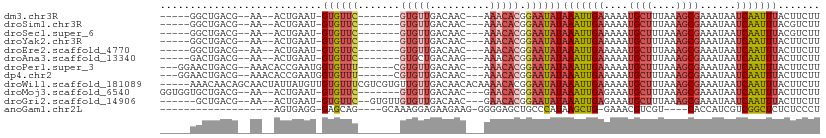
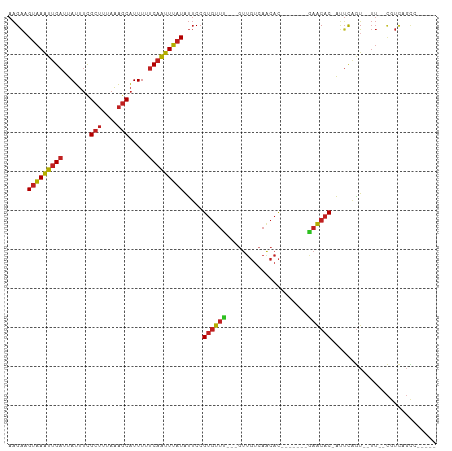
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:39 2011