| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,648,680 – 2,648,766 |
| Length | 86 |
| Max. P | 0.999295 |

| Location | 2,648,680 – 2,648,766 |
|---|---|
| Length | 86 |
| Sequences | 14 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 75.88 |
| Shannon entropy | 0.53859 |
| G+C content | 0.52698 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -27.07 |
| Energy contribution | -25.95 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
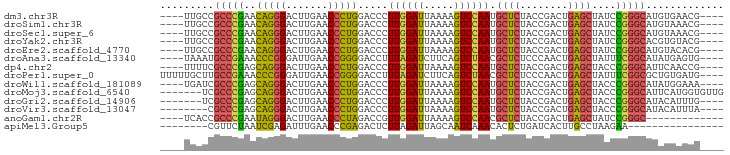
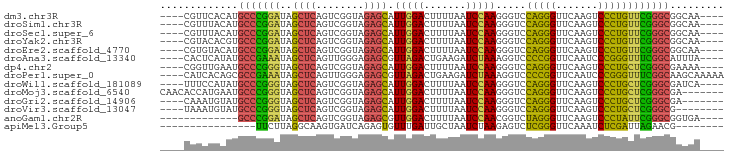
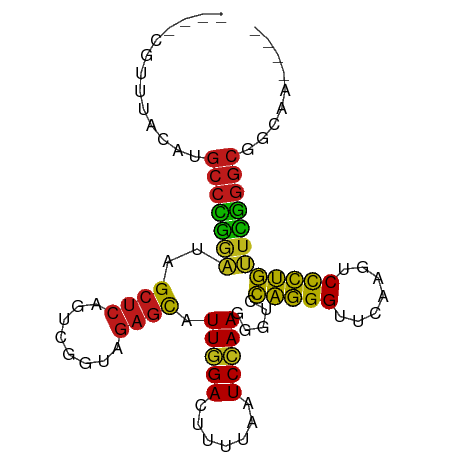
>dm3.chr3R 2648680 86 + 27905053 ----UUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUGAACG---- ----.....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.40, z-score = -3.45, R) >droSim1.chr3R 2679178 86 + 27517382 ----UUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUAAACG---- ----((((.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..))))...---- ( -29.90, z-score = -3.79, R) >droSec1.super_6 2739920 86 + 4358794 ----UUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUAAACG---- ----((((.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..))))...---- ( -29.90, z-score = -3.79, R) >droYak2.chr3R 21067591 86 - 28832112 ----UUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCACGUGUACG---- ----.....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.40, z-score = -3.04, R) >droEre2.scaffold_4770 2897992 86 + 17746568 ----UUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUACACG---- ----.....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.40, z-score = -3.49, R) >droAna3.scaffold_13340 5748475 86 - 23697760 ----UAAAUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAUAUGAGUG---- ----...((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))).......---- ( -31.60, z-score = -4.13, R) >dp4.chr2 11072426 86 - 30794189 ----UUUUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUCAACCG---- ----.....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.10, z-score = -3.23, R) >droPer1.super_0 4793981 90 + 11822988 UUUUUGCUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCGCUGUGAUG---- ...(..(.(((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))..)..)..---- ( -31.40, z-score = -3.36, R) >droWil1.scaffold_181089 12026569 86 + 12369635 ----UGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUAUGGAAA---- ----.....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.10, z-score = -2.85, R) >droMoj3.scaffold_6540 17661356 87 - 34148556 -------UCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUCAUGGUGUUG -------..(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))............. ( -29.10, z-score = -1.99, R) >droGri2.scaffold_14906 2746841 83 - 14172833 -------UCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUACAUUUG---- -------..(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.10, z-score = -3.71, R) >droVir3.scaffold_13047 13142600 82 - 19223366 --------CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUACAUUUA---- --------.(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........---- ( -29.10, z-score = -3.77, R) >anoGam1.chr2R 11448095 77 - 62725911 ----UCACCGCCCGAAUAGGGACUUGAACCCUAGACCGUUGGAUUAAAAGUCCAACGCUCUACCGACUGAGCUAUCCGGGC------------- ----.....(((((..(((((.......)))))....(((((((.....)))))))((((........))))....)))))------------- ( -30.00, z-score = -4.46, R) >apiMel3.Group5 6998220 70 - 13386189 --------CGUUCUAAUCGAGAUUUGAACCCGAGACUCUUAGAUUAGCAAUCAAACACUCUGAUCACUUGCCUAAGAA---------------- --------.((((.(((....))).)))).......((((((...((..((((.......))))..))...)))))).---------------- ( -11.20, z-score = -0.71, R) >consensus ____UUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUAAACG____ .........(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))............. (-27.07 = -25.95 + -1.12)
| Location | 2,648,680 – 2,648,766 |
|---|---|
| Length | 86 |
| Sequences | 14 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 75.88 |
| Shannon entropy | 0.53859 |
| G+C content | 0.52698 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -27.64 |
| Energy contribution | -25.93 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.57 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
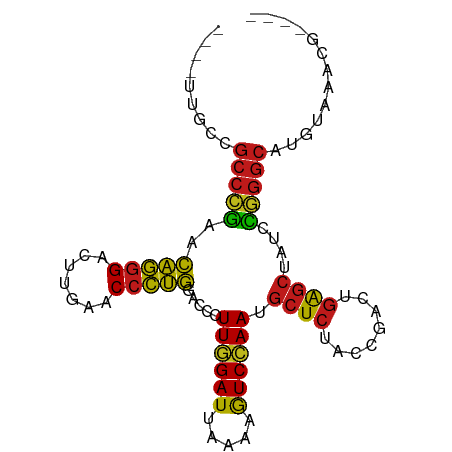
>dm3.chr3R 2648680 86 - 27905053 ----CGUUCACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAA---- ----........((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))....---- ( -33.60, z-score = -3.15, R) >droSim1.chr3R 2679178 86 - 27517382 ----CGUUUACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAA---- ----........((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))....---- ( -33.60, z-score = -3.19, R) >droSec1.super_6 2739920 86 - 4358794 ----CGUUUACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAA---- ----........((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))....---- ( -33.60, z-score = -3.19, R) >droYak2.chr3R 21067591 86 + 28832112 ----CGUACACGUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAA---- ----.......(((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).))..---- ( -33.70, z-score = -2.82, R) >droEre2.scaffold_4770 2897992 86 - 17746568 ----CGUGUACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAA---- ----..(((...((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))))).---- ( -33.90, z-score = -2.96, R) >droAna3.scaffold_13340 5748475 86 + 23697760 ----CACUCAUAUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAUUUA---- ----.......(((((((((..((((........))))...........((((....))))(((((.......))))))))))))))...---- ( -28.00, z-score = -2.09, R) >dp4.chr2 11072426 86 + 30794189 ----CGGUUGAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAAAA---- ----........((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))....---- ( -35.50, z-score = -2.97, R) >droPer1.super_0 4793981 90 - 11822988 ----CAUCACAGCGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAAGCAAAAA ----.......(((((((((..((((........))))...........((((....))))(((((.......))))))))))))..))..... ( -27.50, z-score = -1.65, R) >droWil1.scaffold_181089 12026569 86 - 12369635 ----UUUCCAUAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCA---- ----........((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))....---- ( -35.50, z-score = -3.53, R) >droMoj3.scaffold_6540 17661356 87 + 34148556 CAACACCAUGAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGA------- ............((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).------- ( -35.50, z-score = -2.92, R) >droGri2.scaffold_14906 2746841 83 + 14172833 ----CAAAUGUAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGA------- ----........((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).------- ( -35.50, z-score = -3.67, R) >droVir3.scaffold_13047 13142600 82 + 19223366 ----UAAAUGUAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG-------- ----.........(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).-------- ( -35.20, z-score = -4.02, R) >anoGam1.chr2R 11448095 77 + 62725911 -------------GCCCGGAUAGCUCAGUCGGUAGAGCGUUGGACUUUUAAUCCAACGGUCUAGGGUUCAAGUCCCUAUUCGGGCGGUGA---- -------------(((((((((..........((((.(((((((.......))))))).))))(((.......)))))))))))).....---- ( -33.50, z-score = -3.55, R) >apiMel3.Group5 6998220 70 + 13386189 ----------------UUCUUAGGCAAGUGAUCAGAGUGUUUGAUUGCUAAUCUAAGAGUCUCGGGUUCAAAUCUCGAUUAGAACG-------- ----------------((((((((..((..((((((...))))))..))..))))))))..(((((.......)))))........-------- ( -20.00, z-score = -2.33, R) >consensus ____CGUUUACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAA____ .............(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))......... (-27.64 = -25.93 + -1.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:34 2011