| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,648,318 – 2,648,438 |
| Length | 120 |
| Max. P | 0.968541 |

| Location | 2,648,318 – 2,648,438 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Shannon entropy | 0.09350 |
| G+C content | 0.56346 |
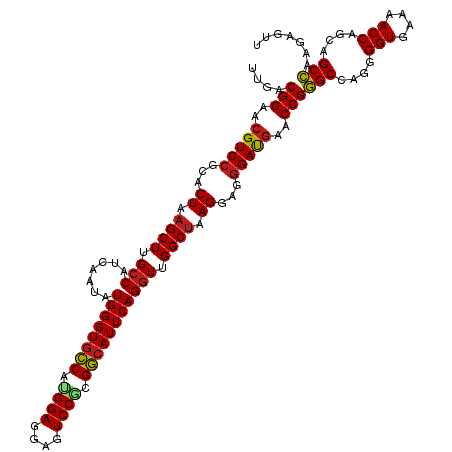
| Mean single sequence MFE | -47.19 |
| Consensus MFE | -42.19 |
| Energy contribution | -43.75 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
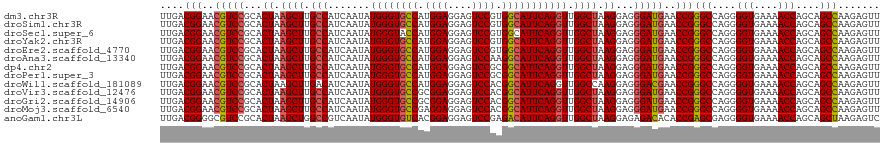
>dm3.chr3R 2648318 120 + 27905053 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -50.11, z-score = -3.17, R) >droSim1.chr3R 2678846 120 + 27517382 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -50.11, z-score = -3.17, R) >droSec1.super_6 2739546 120 + 4358794 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUACCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((.(((((((....))))))).)))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -43.51, z-score = -1.68, R) >droYak2.chr3R 21067177 120 - 28832112 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -50.11, z-score = -3.17, R) >droEre2.scaffold_4770 2897166 120 + 17746568 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -50.11, z-score = -3.17, R) >droAna3.scaffold_13340 5407788 120 + 23697760 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCAAGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -46.21, z-score = -2.43, R) >dp4.chr2 19729335 120 + 30794189 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGCGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -45.51, z-score = -1.82, R) >droPer1.super_3 2465566 120 + 7375914 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGCGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -45.51, z-score = -1.82, R) >droWil1.scaffold_181089 351336 120 - 12369635 UUGACGGAACGUCCGCACUAAGCUUGACAUCAAUAUGGGUGCCAUGGAGGAGUCCACGGCAUUCAGGUUGGCCAAGGAGGGACGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((...((((...(((((.((((((((.((((....)))).)))))))).))))).)))).)))))))..)))(((....(((....)))....)))....... ( -44.40, z-score = -2.06, R) >droVir3.scaffold_12476 5848 120 + 9040 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCGCGGAGGAGUCCACGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((.(((....))).)))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -48.01, z-score = -2.49, R) >droGri2.scaffold_14906 11430746 120 + 14172833 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCGCGGAGGAGUCCACGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((.(((....))).)))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -48.01, z-score = -2.49, R) >droMoj3.scaffold_6540 16492852 120 + 34148556 UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCGAGGAGGAGUCCACGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......(((((((((.(((....))).)))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... ( -47.21, z-score = -2.64, R) >anoGam1.chr3L 27269753 120 + 41284009 UUGACGGGGCGUCCGCACUAAGCUGGCCGUCAAUAUGGGUGUCACGGAGGAGUCCGAGACAUUCAGGUUGGCUAAGGAGAGACACACCGAGCGAGGGGUGAAAACCAGCAGCUAAGAGUC ....(((.(.((((...((.(((..(((.......((((((((.((((....)))).)))))))))))..))).))..).))).).)))(((....(((....)))....)))....... ( -44.71, z-score = -2.41, R) >consensus UUGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGCGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUU ....(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))....... (-42.19 = -43.75 + 1.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:33 2011