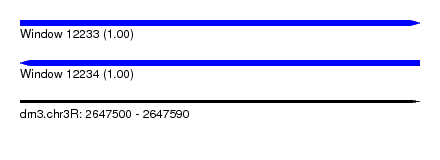
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,647,500 – 2,647,590 |
| Length | 90 |
| Max. P | 0.999411 |

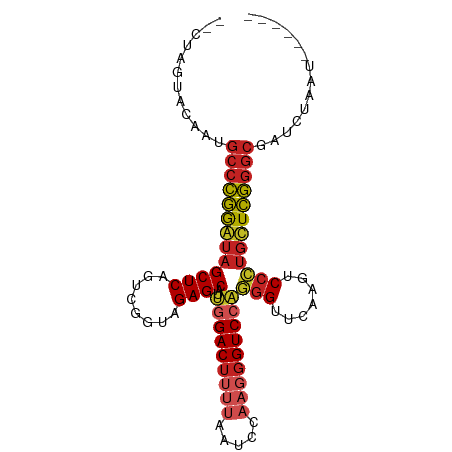
| Location | 2,647,500 – 2,647,590 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.55563 |
| G+C content | 0.53487 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -29.21 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.998759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2647500 90 + 27905053 --CUUUUGCAAGGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGAUCCGCU------ --..........(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))........------ ( -33.30, z-score = -1.99, R) >droSec1.super_14649 292 79 + 644 --CUCGUGCCAAGCGCGGAUAGCUCAGGCGGAAGAGCCUCGGACUUGCACUUCAAGGGUCCGACGAUCAAGUCCGUGCUCG----------------- --.........(((((((((......(((......)))((((((((.........)))))))).......)))))))))..----------------- ( -29.40, z-score = -0.85, R) >droYak2.chr3R 21066731 90 - 28832112 --CUUGUACAAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGAUCUACU------ --...(((.(.((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).).))).------ ( -34.30, z-score = -3.11, R) >droEre2.scaffold_4770 2896722 90 + 17746568 --CUUGUGCAAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGAUCCACU------ --...(((...((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))...))).------ ( -35.80, z-score = -3.10, R) >droAna3.scaffold_13340 5404190 90 + 23697760 --GCAACCAAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCGACU------ --.........((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).......------ ( -35.50, z-score = -2.68, R) >dp4.chr2 19728801 95 + 30794189 ---CGGUUGAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGACUUAAAUUUUUU ---........((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))............. ( -35.50, z-score = -2.68, R) >droWil1.scaffold_181089 343937 85 - 12369635 -------CAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCAAUU------ -------....((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).......------ ( -35.40, z-score = -3.61, R) >droVir3.scaffold_12476 5274 90 + 9040 --CUAAUCAUAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCUCAU------ --.........((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).......------ ( -35.50, z-score = -3.21, R) >droMoj3.scaffold_6540 16492246 90 + 34148556 --CCGGUAGAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCUUAU------ --.....(((.((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))).)))...------ ( -37.50, z-score = -3.15, R) >droGri2.scaffold_14624 1638368 93 - 4233967 --CUAUCACGGCGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAUGUCCCCGGUUCAAUCCCGGGUUUCGGCAAAAAGUGAAU--- --...((((...(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))).....))))..--- ( -29.40, z-score = -1.86, R) >anoGam1.chr2R 48734959 98 + 62725911 CUUUCGCAACUGGCCCGGAUAGCUCAGUCGGUAGAGCGUUGGACUUUUAAUCCAACGGUCAAGGGUUCAAGUCCCUUUUCGGGCGACAAUAAUGCUUU .....(((..(.(((((((..............((.(((((((.......))))))).))(((((.......)))))))))))).)......)))... ( -31.10, z-score = -1.70, R) >consensus __CUAGUACAAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCUAAU______ ............(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).............. (-29.21 = -29.73 + 0.52)

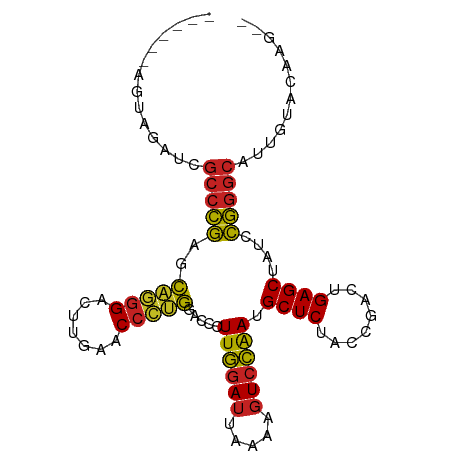
| Location | 2,647,500 – 2,647,590 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.55563 |
| G+C content | 0.53487 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.01 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2647500 90 - 27905053 ------AGCGGAUCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCCUUGCAAAAG-- ------.((((...(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).)))).....-- ( -31.20, z-score = -2.85, R) >droSec1.super_14649 292 79 - 644 -----------------CGAGCACGGACUUGAUCGUCGGACCCUUGAAGUGCAAGUCCGAGGCUCUUCCGCCUGAGCUAUCCGCGCUUGGCACGAG-- -----------------.((((.((((((((....((((....))))....))))))))..))))....(((.((((.......))))))).....-- ( -30.70, z-score = -1.59, R) >droYak2.chr3R 21066731 90 + 28832112 ------AGUAGAUCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUUGUACAAG-- ------.(((.(..(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..).)))...-- ( -31.90, z-score = -3.63, R) >droEre2.scaffold_4770 2896722 90 - 17746568 ------AGUGGAUCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUUGCACAAG-- ------.(((.(..(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..).)))...-- ( -33.30, z-score = -3.56, R) >droAna3.scaffold_13340 5404190 90 - 23697760 ------AGUCGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUUGGUUGC-- ------(..(((..(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..)))..)..-- ( -30.80, z-score = -1.91, R) >dp4.chr2 19728801 95 - 30794189 AAAAAAUUUAAGUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUCAACCG--- ..............(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).........--- ( -29.10, z-score = -2.87, R) >droWil1.scaffold_181089 343937 85 + 12369635 ------AAUUGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUG------- ------........(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).....------- ( -29.10, z-score = -3.52, R) >droVir3.scaffold_12476 5274 90 - 9040 ------AUGAGCUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUAUGAUUAG-- ------........(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..........-- ( -29.10, z-score = -2.71, R) >droMoj3.scaffold_6540 16492246 90 - 34148556 ------AUAAGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUCUACCGG-- ------...(((..(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..))).....-- ( -30.80, z-score = -2.98, R) >droGri2.scaffold_14624 1638368 93 + 4233967 ---AUUCACUUUUUGCCGAAACCCGGGAUUGAACCGGGGACAUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCGCCGUGAUAG-- ---..((((....(((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))..))))...-- ( -32.60, z-score = -3.81, R) >anoGam1.chr2R 48734959 98 - 62725911 AAAGCAUUAUUGUCGCCCGAAAAGGGACUUGAACCCUUGACCGUUGGAUUAAAAGUCCAACGCUCUACCGACUGAGCUAUCCGGGCCAGUUGCGAAAG ...(((..((((..(((((..(((((.......)))))....(((((((.....)))))))((((........))))....))))))))))))..... ( -34.10, z-score = -3.74, R) >consensus ______AGUAGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUUGUACAAG__ ..............(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))............ (-26.32 = -26.01 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:32 2011