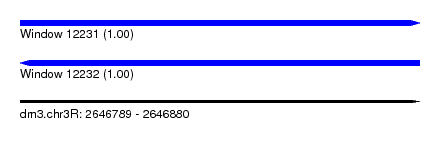
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,646,789 – 2,646,880 |
| Length | 91 |
| Max. P | 0.998849 |

| Location | 2,646,789 – 2,646,880 |
|---|---|
| Length | 91 |
| Sequences | 13 |
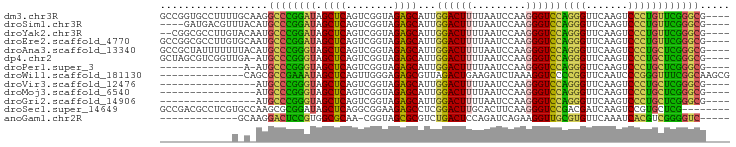
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.74 |
| Shannon entropy | 0.57781 |
| G+C content | 0.57213 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -26.47 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
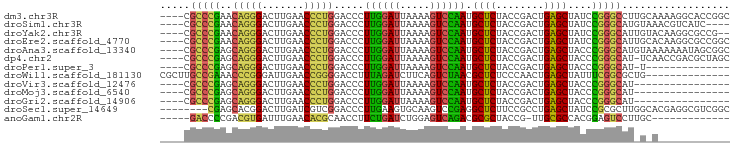
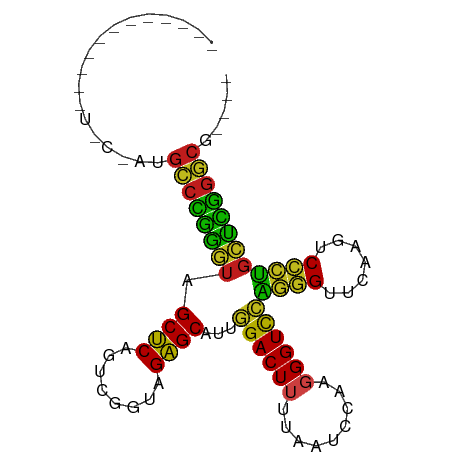
>dm3.chr3R 2646789 91 + 27905053 ----CGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCCUUGCAAAAGGCACCGGC ----........(((((.......))))).....((((((.....)))))).((((........))))...(((((((((....))))).)))). ( -33.10, z-score = -2.81, R) >droSim1.chr3R 2679182 87 + 27517382 ----CGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUAAACGUCAUC---- ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..............---- ( -29.40, z-score = -3.17, R) >droYak2.chr3R 7765298 89 + 28832112 ----CGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUUGUACAAGGCGCCG-- ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))................-- ( -29.40, z-score = -2.21, R) >droEre2.scaffold_4770 2894346 91 + 17746568 ----CGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUUGCACAAGGCGCCGGC ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))...((.....))...... ( -30.30, z-score = -1.56, R) >droAna3.scaffold_13340 18290686 91 - 23697760 ----CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUGUAAAAAAAUAGCGGC ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).................. ( -29.10, z-score = -2.17, R) >dp4.chr2 11065306 90 - 30794189 ----CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAU-UCAACCGACGCUAGC ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..-............... ( -29.10, z-score = -2.41, R) >droPer1.super_3 4910725 76 - 7375914 ----CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAU-U-------------- ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..-.-------------- ( -29.10, z-score = -3.77, R) >droWil1.scaffold_181130 5582661 81 - 16660200 CGCUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCGCUG-------------- .((..((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))..-------------- ( -30.00, z-score = -3.31, R) >droVir3.scaffold_12476 3683 75 - 9040 ----CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAU---------------- ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..---------------- ( -29.10, z-score = -3.83, R) >droMoj3.scaffold_6540 17656227 75 - 34148556 ----CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAU---------------- ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..---------------- ( -29.10, z-score = -3.83, R) >droGri2.scaffold_14906 2742470 75 - 14172833 ----CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAU---------------- ----.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..---------------- ( -29.10, z-score = -3.83, R) >droSec1.super_14649 273 87 - 644 --------CGAGCACGGACUUGAUCGUCGGACCCUUGAAGUGCAAGUCCGAGGCUCUUCCGCCUGAGCUAUCCGCGCUUGGCACGAGGCGUCGGC --------.((((.((((((((....((((....))))....))))))))..))))...((((((.((((........)))).).)))))..... ( -34.10, z-score = -0.46, R) >anoGam1.chr2R 48734135 76 + 62725911 -----GACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCG-UUGCGCCACGGAGUCCUUGC------------- -----(((.(((.((((.......))))......((((((.....)))))).((((.....-..))))..))).))).....------------- ( -21.80, z-score = -1.05, R) >consensus ____CGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAU_G_A____________ .....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).................. (-26.47 = -25.60 + -0.87)
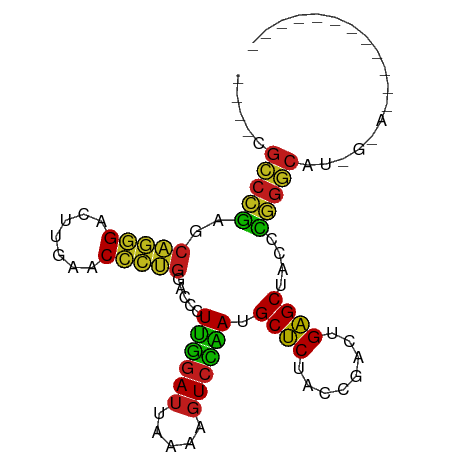
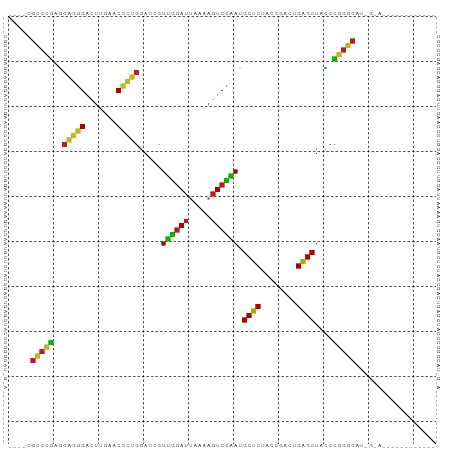
| Location | 2,646,789 – 2,646,880 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.74 |
| Shannon entropy | 0.57781 |
| G+C content | 0.57213 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -29.49 |
| Energy contribution | -28.11 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2646789 91 - 27905053 GCCGGUGCCUUUUGCAAGGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCG---- (((((.(((((....)))))))(((((((((........))))..(((((((((.....)))))))))(((.......)))))))).))).---- ( -35.70, z-score = -2.07, R) >droSim1.chr3R 2679182 87 - 27517382 ----GAUGACGUUUACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCG---- ----..............(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -33.30, z-score = -2.98, R) >droYak2.chr3R 7765298 89 - 28832112 --CGGCGCCUUGUACAAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCG---- --................(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -33.30, z-score = -2.11, R) >droEre2.scaffold_4770 2894346 91 - 17746568 GCCGGCGCCUUGUGCAAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCG---- ....((((...))))...(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -36.60, z-score = -2.19, R) >droAna3.scaffold_13340 18290686 91 + 23697760 GCCGCUAUUUUUUUACAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG---- ..................(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -35.20, z-score = -3.01, R) >dp4.chr2 11065306 90 + 30794189 GCUAGCGUCGGUUGA-AUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG---- ..((((....)))).-..(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -35.70, z-score = -2.23, R) >droPer1.super_3 4910725 76 + 7375914 --------------A-AUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG---- --------------.-..(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -35.20, z-score = -4.19, R) >droWil1.scaffold_181130 5582661 81 + 16660200 --------------CAGCGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAAGCG --------------..(((((((((..((((........))))...........((((....))))(((((.......))))))))))))..)). ( -27.50, z-score = -1.55, R) >droVir3.scaffold_12476 3683 75 + 9040 ----------------AUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG---- ----------------..(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -35.20, z-score = -4.22, R) >droMoj3.scaffold_6540 17656227 75 + 34148556 ----------------AUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG---- ----------------..(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -35.20, z-score = -4.22, R) >droGri2.scaffold_14906 2742470 75 + 14172833 ----------------AUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG---- ----------------..(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).---- ( -35.20, z-score = -4.22, R) >droSec1.super_14649 273 87 + 644 GCCGACGCCUCGUGCCAAGCGCGGAUAGCUCAGGCGGAAGAGCCUCGGACUUGCACUUCAAGGGUCCGACGAUCAAGUCCGUGCUCG-------- (((((....))).))..(((((((((......(((......)))((((((((.........)))))))).......)))))))))..-------- ( -32.50, z-score = -0.38, R) >anoGam1.chr2R 48734135 76 - 62725911 -------------GCAAGGACUCCGUGGCGCAA-CGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUC----- -------------.....(((((((..((((..-.....)))).((((((....).))))).....(((((.......))))))))))))----- ( -24.70, z-score = -0.54, R) >consensus ____________U_C_AUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCG____ ..................((((((((.((((........))))...((((((.........))))))((((.......))))))))))))..... (-29.49 = -28.11 + -1.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:30 2011