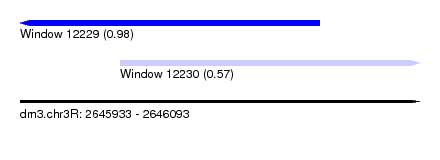
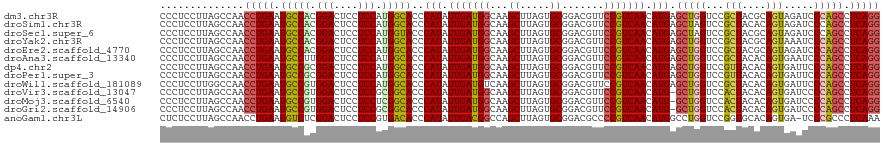
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,645,933 – 2,646,093 |
| Length | 160 |
| Max. P | 0.981552 |

| Location | 2,645,933 – 2,646,053 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.87 |
| Shannon entropy | 0.09676 |
| G+C content | 0.57115 |
| Mean single sequence MFE | -47.28 |
| Consensus MFE | -43.69 |
| Energy contribution | -43.75 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
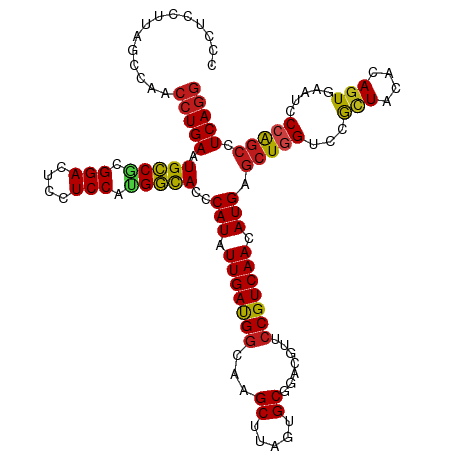
>dm3.chr3R 2645933 120 - 27905053 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -50.11, z-score = -3.23, R) >droSim1.chr3R 24838415 120 + 27517382 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -50.11, z-score = -3.23, R) >droSec1.super_6 2737087 120 - 4358794 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUACCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((.(((((((....))))))).)))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -43.51, z-score = -1.66, R) >droYak2.chr3R 21064716 120 + 28832112 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -50.11, z-score = -3.23, R) >droEre2.scaffold_4770 2893871 120 - 17746568 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGUGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((((((....)))))))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -50.11, z-score = -3.23, R) >droAna3.scaffold_13340 18289851 120 + 23697760 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCAAGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -46.21, z-score = -2.43, R) >dp4.chr2 11064733 120 + 30794189 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGCGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -45.51, z-score = -1.83, R) >droPer1.super_3 4910227 120 + 7375914 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGCGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -45.51, z-score = -1.83, R) >droWil1.scaffold_181089 12025040 120 - 12369635 UGACGGAACGUCCGCACUAAGCUUGACAUCAAUAUGGGUGCCAUGGAGGAGUCCACGGCAUUCAGGUUGGCCAAGGAGGGACGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((...((((...(((((.((((((((.((((....)))).)))))))).))))).)))).)))))))..)))(((....(((....)))....)))........ ( -44.40, z-score = -2.04, R) >droVir3.scaffold_13047 13141982 120 + 19223366 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCGCGGAGGAGUCCACGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((.(((....))).)))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -48.01, z-score = -2.49, R) >droMoj3.scaffold_6540 17655583 120 + 34148556 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCGAGGAGGAGUCCACGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((.(((....))).)))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -47.21, z-score = -2.60, R) >droGri2.scaffold_14906 2741966 120 + 14172833 UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCGCGGAGGAGUCCACGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......(((((((((.(((....))).)))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ ( -48.01, z-score = -2.49, R) >anoGam1.chr3L 13644215 120 + 41284009 UGACGGGGCGUCCGCACUAAGCUGGCCGUCAAUAUGGGUGUCACGGAGGAGUCCGAGACAUUCAGGUUGGCUAAGGAGAGACACACCGAGCGAGGGGUGAAAACCAGCAGCUAAGAGUCU .(((((.(.((((...((.(((..(((.......((((((((.((((....)))).)))))))))))..))).))..).))).).)).(((....(((....)))....)))....))). ( -45.81, z-score = -2.44, R) >consensus UGACGGAACGUCCGCACUAAGCUUGCCAUCAAUAUGGGUGCCAUGGAGGAGUCCGCGGCAUUCAGGUUGGCUAAGGAGGGAUGAACCGGGCCAGGGGUGAAAACCAGCAGCCAAGAGUUC ...(((..(((((...((.((((.(((.......((((((((.((((....)))).))))))))))).)))).))...)))))..)))(((....(((....)))....)))........ (-43.69 = -43.75 + 0.05)
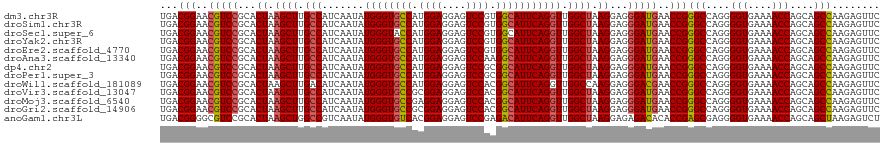
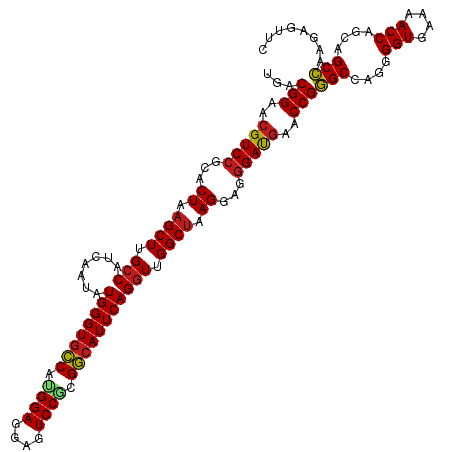
| Location | 2,645,973 – 2,646,093 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Shannon entropy | 0.15806 |
| G+C content | 0.57266 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.13 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2645973 120 + 27905053 CCCUCCUUAGCCAACCUGAAUGCCACGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGCUACGCAGUAGAUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))...........(((.((((((.(((((.....))).))...))))))((((..((....))..))))...)))))))) ( -39.00, z-score = -1.92, R) >droSim1.chr3R 24838455 120 - 27517382 CCCUCCUUAGCCAACCUGAAUGCCACGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUAGUCCGCUACACAGUAGAUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))...........(((.((((((.(((((.....))).))...)))))).(((..((....))..)))....)))))))) ( -36.60, z-score = -2.15, R) >droSec1.super_6 2737127 120 + 4358794 CCCUCCUUAGCCAACCUGAAUGCCACGGACUCCUCCAUGGUACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUAGUCCGCUACGCAGUAGAUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))...........(((.((((((.(((((.....))).))...)))))).(((..((....))..)))....)))))))) ( -34.60, z-score = -1.35, R) >droYak2.chr3R 21064756 120 - 28832112 CCCUCCUUAGCCAACCUGAAUGCCACGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGCUACGCAGUAAAUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))....(((((.((((.((((((.(((((.....))).))...)))))).....))))..)))))..........))))) ( -35.00, z-score = -1.13, R) >droEre2.scaffold_4770 2893911 120 + 17746568 CCCUCCUUAGCCAACCUGAAUGCCACGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGCUACGCAGUAGAUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))...........(((.((((((.(((((.....))).))...))))))((((..((....))..))))...)))))))) ( -39.00, z-score = -1.92, R) >droAna3.scaffold_13340 18289891 120 - 23697760 CCCUCCUUAGCCAACCUGAAUGCCUUGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGCUACACAGUGAAUCCCAGCCUCAGG ..............(((((.((((.((((....)))).))))...........(((.((((((.(((((.....))).))...))))))(((.((((....)))).)))...)))))))) ( -37.50, z-score = -1.74, R) >dp4.chr2 11064773 120 - 30794189 CCCUCCUUAGCCAACCUGAAUGCCGCGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGUUACACAGUGAUUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))...........(((.((((((.(((((.....))).))...))))))((...(((((...)))))..)).)))))))) ( -35.70, z-score = -1.05, R) >droPer1.super_3 4910267 120 - 7375914 CCCUCCUUAGCCAACCUGAAUGCCGCGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGUUACACAGUGAUUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))...........(((.((((((.(((((.....))).))...))))))((...(((((...)))))..)).)))))))) ( -35.70, z-score = -1.05, R) >droWil1.scaffold_181089 12025080 120 + 12369635 CCCUCCUUGGCCAACCUGAAUGCCGUGGACUCCUCCAUGGCACCCAUAUUGAUGUCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGCUACACAGUGAUUCCCAGCCUCAGG .(((.(((((((((..((..(((((((((....)))))))))..))..)))..))))))....(((..((((...))))..)))(((((((..((((....))))....)))).)))))) ( -39.90, z-score = -2.13, R) >droVir3.scaffold_13047 13142022 119 - 19223366 CCCUCCUUAGCCAACCUGAAUGCCGUGGACUCCUCCGCGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUG-GCUGGUCCACUACACAGUGAUCCCCAGCCUCAGG ..............(((((.(((((((((....))))))))).......((.((((..((.....))((.....)))))).)).(-(((((..((((....))))....))))))))))) ( -43.30, z-score = -2.94, R) >droMoj3.scaffold_6540 17655623 119 - 34148556 CCCUCCUUAGCCAACCUGAAUGCCGUGGACUCCUCCUCGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUG-GCUGGUCCACUACACAGUGAUCCCCAGCCUCAGG ..............(((((.(((((.(((....))).))))).......((.((((..((.....))((.....)))))).)).(-(((((..((((....))))....))))))))))) ( -39.70, z-score = -2.14, R) >droGri2.scaffold_14906 2742006 119 - 14172833 CCCUCCUUAGCCAACCUGAAUGCCGUGGACUCCUCCGCGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUG-GCUGGUCCACUACACAGUGAUCCCCAGCCUCAGG ..............(((((.(((((((((....))))))))).......((.((((..((.....))((.....)))))).)).(-(((((..((((....))))....))))))))))) ( -43.30, z-score = -2.94, R) >anoGam1.chr3L 13644255 119 - 41284009 CUCUCCUUAGCCAACCUGAAUGUCUCGGACUCCUCCGUGACACCCAUAUUGACGGCCAGCUUAGUGCGGACGCCCCGUCAACAUGGCCUGGUCCGGUGCACAGUGA-UCCCGCCCUCAAA .........(((.(((....((((.((((....)))).)))).((((.(((((((((.((.....)))).....))))))).))))...)))..))).....(((.-...)))....... ( -33.20, z-score = -0.21, R) >consensus CCCUCCUUAGCCAACCUGAAUGCCGCGGACUCCUCCAUGGCACCCAUAUUGAUGGCAAGCUUAGUGCGGACGUUCCGUCAACAUGAGCUGGUCCGCUACACAGUGAAUCCCAGCCUCAGG ..............(((((.(((((.(((....))).)))))..(((.(((((((...((.....)).......))))))).))).(((((...(((....))).....))))).))))) (-30.33 = -30.13 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:29 2011