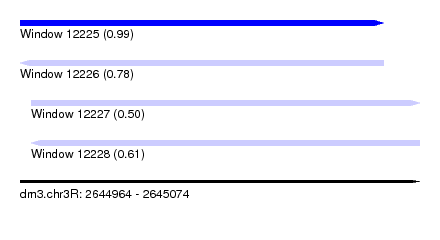
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,644,964 – 2,645,074 |
| Length | 110 |
| Max. P | 0.993401 |

| Location | 2,644,964 – 2,645,064 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 63.92 |
| Shannon entropy | 0.70286 |
| G+C content | 0.43501 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -10.14 |
| Energy contribution | -9.14 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
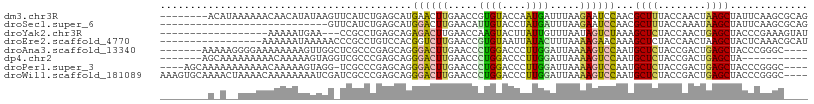
>dm3.chr3R 2644964 100 + 27905053 --------ACAUAAAAAACAACAUAUAAGUUCAUCUGAGCAUGAACUUGAACCGUGUACCAAUGAUUUAAGAAUCCAACGCUUUACCAACUAAGCUAUUCAAGCGCAG --------..........................(((.((.((((((((((.(((......))).))))))........((((........))))..)))).)).))) ( -16.40, z-score = -1.99, R) >droSec1.super_6 2736077 80 + 4358794 ----------------------------GUUCAUCUGAGCAUGGACUUGAACAUUGUACCUAUGAUUUAAGAAUCCAACGCUUUACCAAAUAAGCUAUUCAAGCGCAG ----------------------------......(((.((.(((((((((((((.......))).))))))..))))..((((........)))).......)).))) ( -16.00, z-score = -1.74, R) >droYak2.chr3R 21063617 87 - 28832112 ------------------AAAAAUGAA---CCGCCUGAGCAGAGACUUGAACCAAGUACUUAUUGUUUAAUAGUCUAAAGCUCUACCAACUGAGCUACCCGAAAGUAU ------------------.........---.....((((((((((((((...))))).)))..)))))).........(((((........)))))...(....)... ( -15.10, z-score = -1.73, R) >droEre2.scaffold_4770 2892350 91 + 17746568 -----------------AAAAAAUAAAAACCCGCCUGUCCACGGUCUUGAACCGUGUAAUUAUACUUUAAAAGAACAAAGCUCUACCAACUAAGCUACUCAAACGCAU -----------------...............((.....((((((.....))))))......................((((..........))))........)).. ( -11.40, z-score = -1.41, R) >droAna3.scaffold_13340 18289467 97 - 23697760 -------AAAAAGGGGAAAAAAAAGUUGGCUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC---- -------....((((.......(((((.((((....))))...)))))...))))...(((((((((.....)))))).((((........)))).....))).---- ( -29.90, z-score = -0.92, R) >dp4.chr2 11064283 90 - 30794189 -------AGCAAAAAAAAACAAAAAGUAGGUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUA----------- -------.................(((.(((((...((((((((.((........)).)))((((((.....)))))))))))...)))))..))).----------- ( -25.50, z-score = -2.30, R) >droPer1.super_3 4897978 99 - 7375914 ----AGCAAAAAAAAAAACAAAAAGUAGG-UCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC---- ----.........................-..(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))---- ( -27.40, z-score = -1.47, R) >droWil1.scaffold_181089 12017435 104 + 12369635 AAAGUGCAAAACUAAAACAAAAAAAAUCGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC---- ..(((.....)))...................(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))---- ( -28.00, z-score = -2.52, R) >consensus ________A_A__AAAAAAAAAAAAAAAG_UCGCCCGAGCAGGGACUUGAACCCUGGACCCAUGGAUUAAAAGUCCAAUGCUCUACCAACUGAGCUACCCGAGC____ ..........................................((((((.....((((....)))).....))))))...((((........))))............. (-10.14 = -9.14 + -1.00)

| Location | 2,644,964 – 2,645,064 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 63.92 |
| Shannon entropy | 0.70286 |
| G+C content | 0.43501 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
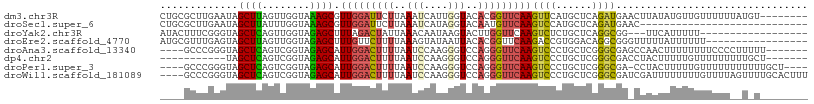
>dm3.chr3R 2644964 100 - 27905053 CUGCGCUUGAAUAGCUUAGUUGGUAAAGCGUUGGAUUCUUAAAUCAUUGGUACACGGUUCAAGUUCAUGCUCAGAUGAACUUAUAUGUUGUUUUUUAUGU-------- (.((((((...((((...))))...)))))).)...........(((....(((..((..((((((((......))))))))..))..))).....))).-------- ( -18.10, z-score = 0.66, R) >droSec1.super_6 2736077 80 - 4358794 CUGCGCUUGAAUAGCUUAUUUGGUAAAGCGUUGGAUUCUUAAAUCAUAGGUACAAUGUUCAAGUCCAUGCUCAGAUGAAC---------------------------- .((.((((((((.((((........))))((((...(((........)))..)))))))))))).)).............---------------------------- ( -15.00, z-score = 0.12, R) >droYak2.chr3R 21063617 87 + 28832112 AUACUUUCGGGUAGCUCAGUUGGUAGAGCUUUAGACUAUUAAACAAUAAGUACUUGGUUCAAGUCUCUGCUCAGGCGG---UUCAUUUUU------------------ ........(..(.(((....((((((((.(((..((((....((.....))...))))..))).)))))).))))).)---..)......------------------ ( -17.00, z-score = -0.18, R) >droEre2.scaffold_4770 2892350 91 - 17746568 AUGCGUUUGAGUAGCUUAGUUGGUAGAGCUUUGUUCUUUUAAAGUAUAAUUACACGGUUCAAGACCGUGGACAGGCGGGUUUUUAUUUUUU----------------- ((.((((((.......((((((.....((((((......)))))).))))))(((((((...)))))))..)))))).))...........----------------- ( -20.40, z-score = -1.16, R) >droAna3.scaffold_13340 18289467 97 + 23697760 ----GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAACUUUUUUUUCCCCUUUUU------- ----(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).........................------- ( -35.20, z-score = -2.30, R) >dp4.chr2 11064283 90 + 30794189 -----------UAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGACCUACUUUUUGUUUUUUUUUGCU------- -----------.(((.(((..(((((((((.(((((((((.....)))))))))(((.......))))))))((....))))))..))).........)))------- ( -27.00, z-score = -1.45, R) >droPer1.super_3 4897978 99 + 7375914 ----GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGA-CCUACUUUUUGUUUUUUUUUUUGCU---- ----(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))..-.........................---- ( -35.20, z-score = -2.51, R) >droWil1.scaffold_181089 12017435 104 - 12369635 ----GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCGAUUUUUUUUGUUUUAGUUUUGCACUUU ----(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))................................ ( -35.20, z-score = -2.64, R) >consensus ____GCUCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAACCAAGGGUACAGGGUUCAAGUCCCUGCUCAGGCGA_CUUAUUUUUUUUUUUU__U_U________ .............((((........)))).(((((((((..(((....)))..)))))))))((((......))))................................ (-10.14 = -10.70 + 0.56)
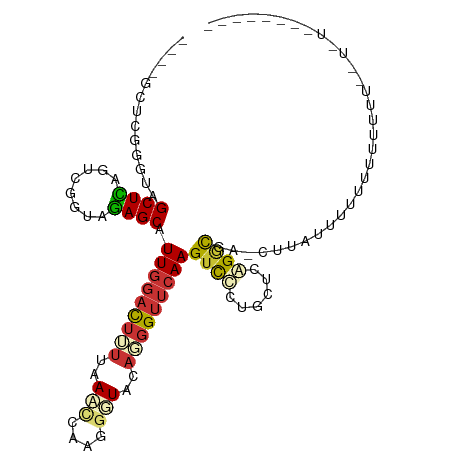
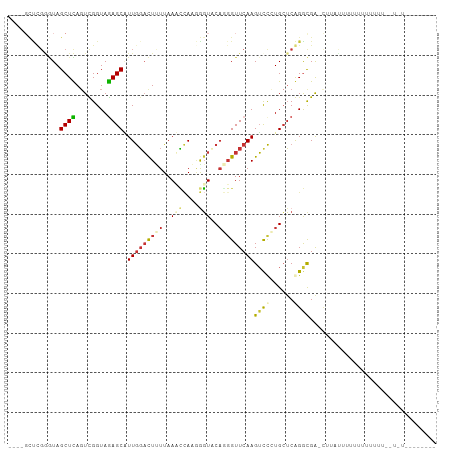
| Location | 2,644,967 – 2,645,074 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 60.48 |
| Shannon entropy | 0.79269 |
| G+C content | 0.44371 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -4.64 |
| Energy contribution | -4.38 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
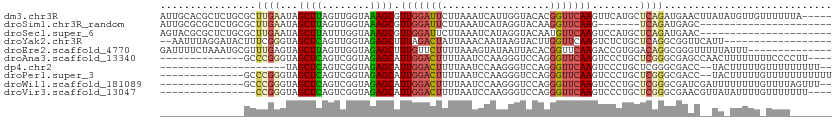
>dm3.chr3R 2644967 107 + 27905053 -----UAAAAAACAACAUAUAAGUUCAUCUGAGCAUGAACUUGAACCGUGUACCAAUGAUUUAAGAAUCCAACGCUUUACCAACUAAGCUAUUCAAGCGCAGAGCGUGCAAU -----.................((...((((.((.((((((((((.(((......))).))))))........((((........))))..)))).)).))))....))... ( -20.80, z-score = -1.87, R) >droSim1.chr3R_random 236029 83 + 1307089 ----------------------GCUCAUCUGA-------CUUGAACCUUGUACCUAUGAUUUAAGAAUCCAACGCUUUACCAACUAAGCUAUUCAAGCGCAGAGCGCGCAAU ----------------------((.(.((((.-------((((((............((((....))))....((((........))))..))))))..))))..).))... ( -16.00, z-score = -1.27, R) >droSec1.super_6 2736077 90 + 4358794 ----------------------GUUCAUCUGAGCAUGGACUUGAACAUUGUACCUAUGAUUUAAGAAUCCAACGCUUUACCAAAUAAGCUAUUCAAGCGCAGAGCGCGUACU ----------------------((...((((.((.(((((((((((((.......))).))))))..))))..((((........)))).......)).))))..))..... ( -19.90, z-score = -1.41, R) >droYak2.chr3R 21063620 92 - 28832112 ------------------AAUGAACCGCCUGAGCAGAGACUUGAACCAAGUACUUAUUGUUUAAUAGUCUAAAGCUCUACCAACUGAGCUACCCGAAAGUAUCCUAAAUU-- ------------------...........((((((((((((((...))))).)))..)))))).(((..((.(((((........)))))...(....)))..)))....-- ( -15.70, z-score = -1.62, R) >droEre2.scaffold_4770 2892353 98 + 17746568 --------------AAAUAAAAACCCGCCUGUCCACGGUCUUGAACCGUGUAAUUAUACUUUAAAAGAACAAAGCUCUACCAACUAAGCUACUCAAACGCAUUUAGAAAAUC --------------............((.....((((((.....))))))......................((((..........))))........))............ ( -11.40, z-score = -0.71, R) >droAna3.scaffold_13340 18289470 94 - 23697760 ----AAGGGGAAAAAAAAGUUGGCUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC-------------- ----.((((.......(((((.((((....))))...)))))...))))...(((((((((.....)))))).((((........)))).....))).-------------- ( -29.90, z-score = -0.80, R) >dp4.chr2 11064286 87 - 30794189 --AAAAAAAAACAAAAAGUA--GGUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUA--------------------- --..............(((.--(((((...((((((((.((........)).)))((((((.....)))))))))))...)))))..))).--------------------- ( -25.50, z-score = -2.81, R) >droPer1.super_3 4897981 96 - 7375914 AAAAAAAAAAACAAAAAGUA--GGUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC-------------- ....................--....(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))-------------- ( -27.40, z-score = -1.86, R) >droWil1.scaffold_181089 12017443 96 + 12369635 --AAACUAAAACAAAAAAAAUCGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGC-------------- --........................(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))-------------- ( -27.40, z-score = -3.06, R) >droVir3.scaffold_13047 13141548 92 - 19223366 ----AAAAAAACAAAAUAUAACGUUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGG---------------- ----..................((((.(((.....)))....)))).((((....((((((.....)))))).((((........))))...))))---------------- ( -22.30, z-score = -2.19, R) >consensus _______AAAA_AAAAA_AA__GCUCGCCCGAGCAGGGACUUGAACCCUGGACCCAUGGAUUAAAAGUCCAAUGCUCUACCAACUGAGCUACCCGAGC______________ .........................................................................((((........))))....................... ( -4.64 = -4.38 + -0.26)



| Location | 2,644,967 – 2,645,074 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 60.48 |
| Shannon entropy | 0.79269 |
| G+C content | 0.44371 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -8.47 |
| Energy contribution | -8.14 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
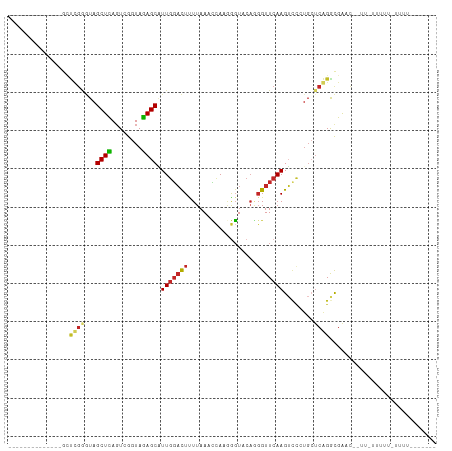
>dm3.chr3R 2644967 107 - 27905053 AUUGCACGCUCUGCGCUUGAAUAGCUUAGUUGGUAAAGCGUUGGAUUCUUAAAUCAUUGGUACACGGUUCAAGUUCAUGCUCAGAUGAACUUAUAUGUUGUUUUUUA----- .......((((.((((((...((((...))))...)))))).)((((....))))...)))(((..((..((((((((......))))))))..))..)))......----- ( -18.00, z-score = 1.65, R) >droSim1.chr3R_random 236029 83 - 1307089 AUUGCGCGCUCUGCGCUUGAAUAGCUUAGUUGGUAAAGCGUUGGAUUCUUAAAUCAUAGGUACAAGGUUCAAG-------UCAGAUGAGC---------------------- ...((.((.((((.((((((((.((((........)))).(((...(((........)))..))).)))))))-------))))))).))---------------------- ( -19.40, z-score = 0.18, R) >droSec1.super_6 2736077 90 - 4358794 AGUACGCGCUCUGCGCUUGAAUAGCUUAUUUGGUAAAGCGUUGGAUUCUUAAAUCAUAGGUACAAUGUUCAAGUCCAUGCUCAGAUGAAC---------------------- .....(.((..((.((((((((.((((........))))((((...(((........)))..)))))))))))).)).)).)........---------------------- ( -17.70, z-score = 0.61, R) >droYak2.chr3R 21063620 92 + 28832112 --AAUUUAGGAUACUUUCGGGUAGCUCAGUUGGUAGAGCUUUAGACUAUUAAACAAUAAGUACUUGGUUCAAGUCUCUGCUCAGGCGGUUCAUU------------------ --......(((....)))(..(.(((....((((((((.(((..((((....((.....))...))))..))).)))))).))))).)..)...------------------ ( -17.20, z-score = 0.42, R) >droEre2.scaffold_4770 2892353 98 - 17746568 GAUUUUCUAAAUGCGUUUGAGUAGCUUAGUUGGUAGAGCUUUGUUCUUUUAAAGUAUAAUUACACGGUUCAAGACCGUGGACAGGCGGGUUUUUAUUU-------------- ........((((.((((((.......((((((.....((((((......)))))).))))))(((((((...)))))))..)))))).))))......-------------- ( -22.40, z-score = -1.18, R) >droAna3.scaffold_13340 18289470 94 + 23697760 --------------GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAACUUUUUUUUCCCCUU---- --------------(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))......................---- ( -35.20, z-score = -2.22, R) >dp4.chr2 11064286 87 + 30794189 ---------------------UAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGACC--UACUUUUUGUUUUUUUUU-- ---------------------.....(((..(((((((((.(((((((((.....)))))))))(((.......))))))))((....))--))))..))).........-- ( -26.60, z-score = -1.87, R) >droPer1.super_3 4897981 96 + 7375914 --------------GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGACC--UACUUUUUGUUUUUUUUUUU --------------(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))....--.................... ( -35.20, z-score = -3.05, R) >droWil1.scaffold_181089 12017443 96 - 12369635 --------------GCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCGAUUUUUUUUGUUUUAGUUU-- --------------(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))........................-- ( -35.20, z-score = -3.28, R) >droVir3.scaffold_13047 13141548 92 + 19223366 ----------------CCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAACGUUAUAUUUUGUUUUUUU---- ----------------(((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))..(((((........)))))....---- ( -29.30, z-score = -2.23, R) >consensus ______________GCUCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAACCAAGGGUACAGGGUUCAAGUCCCUGCUCAGGCGAAC__UU_UUUUU_UUUU_______ ................((((...((((........)))).(((((((..................)))))))........))))............................ ( -8.47 = -8.14 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:27 2011