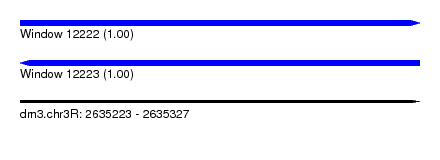
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,635,223 – 2,635,327 |
| Length | 104 |
| Max. P | 0.999378 |

| Location | 2,635,223 – 2,635,327 |
|---|---|
| Length | 104 |
| Sequences | 15 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.48407 |
| G+C content | 0.41259 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.82 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2635223 104 + 27905053 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACAGUCG--- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..)).((((........))))..--- ( -28.07, z-score = -2.73, R) >droEre2.scaffold_4770 2882565 103 + 17746568 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUAGAGCAAACUGAUGAGGAAC-GUCG--- ............(((.((.((((.((((((((((........-..........)))))))))).)))).))))).(((((....((......))....))-))).--- ( -26.07, z-score = -2.51, R) >droYak2.chr3R 21054027 103 - 28832112 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAA-CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAAGUGAUGAGGAACGUCA---- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..))...(((((.....)))))---- ( -28.77, z-score = -2.92, R) >droSec1.super_6 2726444 104 + 4358794 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACAGUCG--- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..)).((((........))))..--- ( -28.07, z-score = -2.73, R) >droSim1.chr3R 2666355 104 + 27517382 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGAUGAGGAACAGUCG--- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..)).((((........))))..--- ( -28.07, z-score = -2.73, R) >dp4.chr2 11054608 107 - 30794189 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAAUGUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUUGAGAACCAAAAAAACUG ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..))...................... ( -26.27, z-score = -3.42, R) >droPer1.super_3 4888140 107 - 7375914 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAAUGUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUUGAGAACCAAAAAAACUG ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..))...................... ( -26.27, z-score = -3.42, R) >droAna3.scaffold_13340 18280312 106 - 23697760 UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAA-CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGGAUAGGAGCACAGCCG- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..))..(((..........)))...- ( -27.27, z-score = -2.54, R) >droWil1.scaffold_181089 12007330 88 + 12369635 UGAACCACUUAAACCUCUCUAGAACCGAAUUUGUCCUUAAAAACUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUAGAACAAA-------------------- ............(((.((.((((.((((((((((...................)))))))))).)))).)))))..............-------------------- ( -19.81, z-score = -2.81, R) >droVir3.scaffold_13047 13131222 87 - 19223366 UGAGCCACAGAAACCUCUCUAGAACCGAAUUUGUCCUUAAAA-CUAGUGUACAACAAAUUCGGAUCUACAGGGUGGACGUGGAACAAA-------------------- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..))..-------------------- ( -27.37, z-score = -3.42, R) >droMoj3.scaffold_6540 17645564 91 - 34148556 UGAGCCACAUAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAUGGUACAACAAAUUCGGAUCUACAGGGUAGACGUGGAACAGAGUGG---------------- ((..((((.(..(((.((.((((.((((((((((((......-....))....)))))))))).)))).)))))..).))))..))......---------------- ( -28.10, z-score = -3.03, R) >droGri2.scaffold_14906 2729190 91 - 14172833 UGAGCCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG-CUAGUGUACAACAAAUUCGGAUCUACAGGGUAGACGUGGAACAAAGUGA---------------- ((..((((....(((.((.((((.((((((((((........-..........)))))))))).)))).)))))....))))..))......---------------- ( -26.27, z-score = -2.69, R) >anoGam1.chr2R 61507584 105 + 62725911 UGCCCCACACAAACCUCUCUAGAACCGAAUUUGUUAUUAAUA--UAAAUUCAAACAAAUUCGGAUCUACAGGGUGGAUGUAGAACAUAAAUCGACGAAGAUAAAUCG- ........(((..(((((.((((.(((((((((((.......--........))))))))))).)))).)))).)..))).........(((......)))......- ( -24.36, z-score = -3.19, R) >apiMel3.Group16 2947300 86 + 5631066 -GCACCACACAAACCUCUCUAGAACCGAAUUUGUCGCCUCU---UA---UCAAACAAAUUCGGAUCUACAGGGUAGAUGUAGAGCAUUAACUG--------------- -((.(.(((...(((.((.((((.((((((((((.......---..---....)))))))))).)))).)))))...))).).))........--------------- ( -24.22, z-score = -4.34, R) >triCas2.ChLG2 9946467 80 - 12900155 UGCACCACACAAACCUCUCUAGAACCGAAUUUGUCGCC------UGAUUUCAAACAAAUUCGGAUCUACAGGGUAGAUGUAGAGCA---------------------- .((.(.(((...(((.((.((((.((((((((((....------((....)).)))))))))).)))).)))))...))).).)).---------------------- ( -24.70, z-score = -3.89, R) >consensus UGAACCACACAAACCUCUCUAGAACCGAAUUUGUCCUUAAAG_CUAGUAUAAAACAAAUUCGGAUCUACAGGGUAGACGUGGAGCAAACUGA_GAG_AA_________ ............(((.((.((((.((((((((((...................)))))))))).)))).))))).................................. (-19.82 = -19.82 + -0.00)

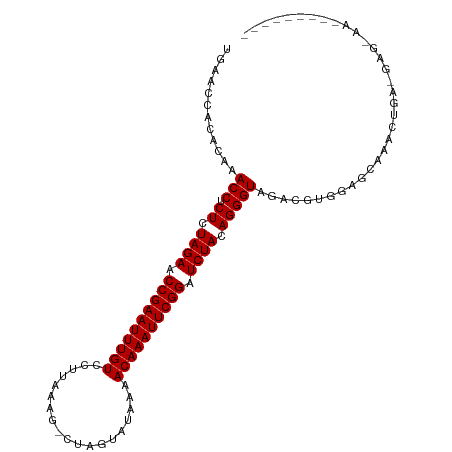
| Location | 2,635,223 – 2,635,327 |
|---|---|
| Length | 104 |
| Sequences | 15 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.48407 |
| G+C content | 0.41259 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -31.12 |
| Energy contribution | -30.72 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.55 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2635223 104 - 27905053 ---CGACUGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA ---.(((((........)))))((..(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...)))))..)) ( -36.00, z-score = -4.56, R) >droEre2.scaffold_4770 2882565 103 - 17746568 ---CGAC-GUUCCUCAUCAGUUUGCUCUACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA ---.(((-((....((......))....))))).(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))............ ( -30.70, z-score = -2.89, R) >droYak2.chr3R 21054027 103 + 28832112 ----UGACGUUCCUCAUCACUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG-UUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA ----..................((..(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...)))))..)) ( -32.50, z-score = -3.66, R) >droSec1.super_6 2726444 104 - 4358794 ---CGACUGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA ---.(((((........)))))((..(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...)))))..)) ( -36.00, z-score = -4.56, R) >droSim1.chr3R 2666355 104 - 27517382 ---CGACUGUUCCUCAUCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA ---.(((((........)))))((..(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...)))))..)) ( -36.00, z-score = -4.56, R) >dp4.chr2 11054608 107 + 30794189 CAGUUUUUUUGGUUCUCAAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUACAUUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA .(((...((((.....))))...)))(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...))))).... ( -33.40, z-score = -4.02, R) >droPer1.super_3 4888140 107 + 7375914 CAGUUUUUUUGGUUCUCAAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUACAUUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA .(((...((((.....))))...)))(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...))))).... ( -33.40, z-score = -4.02, R) >droAna3.scaffold_13340 18280312 106 + 23697760 -CGGCUGUGCUCCUAUCCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG-UUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA -.(((...(((.......)))..)))(((((...(((((.((((.(((((((((((((.......-.....))))))))))))).)))).)).)))...))))).... ( -34.30, z-score = -3.54, R) >droWil1.scaffold_181089 12007330 88 - 12369635 --------------------UUUGUUCUACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAGUUUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUAAGUGGUUCA --------------------..((..((((....(((((.((((.(((((((((((((.............))))))))))))).)))).)).)))....))))..)) ( -31.02, z-score = -5.25, R) >droVir3.scaffold_13047 13131222 87 + 19223366 --------------------UUUGUUCCACGUCCACCCUGUAGAUCCGAAUUUGUUGUACACUAG-UUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUCUGUGGCUCA --------------------..((..(((((...(((((.((((.(((((((((((.(.......-.....).))))))))))).)))).)).)))...)))))..)) ( -31.10, z-score = -4.26, R) >droMoj3.scaffold_6540 17645564 91 + 34148556 ----------------CCACUCUGUUCCACGUCUACCCUGUAGAUCCGAAUUUGUUGUACCAUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUAUGUGGCUCA ----------------.......(..((((((..(((((.((((.(((((((((((.(.......-.....).))))))))))).)))).)).)))..))))))..). ( -34.20, z-score = -5.01, R) >droGri2.scaffold_14906 2729190 91 + 14172833 ----------------UCACUUUGUUCCACGUCUACCCUGUAGAUCCGAAUUUGUUGUACACUAG-CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGCUCA ----------------......((..(((((...(((((.((((.(((((((((((.(.......-.....).))))))))))).)))).)).)))...)))))..)) ( -31.80, z-score = -3.85, R) >anoGam1.chr2R 61507584 105 - 62725911 -CGAUUUAUCUUCGUCGAUUUAUGUUCUACAUCCACCCUGUAGAUCCGAAUUUGUUUGAAUUUA--UAUUAAUAACAAAUUCGGUUCUAGAGAGGUUUGUGUGGGGCA -((((........)))).....((((((((((..(((((.((((.(((((((((((..(((...--.)))...))))))))))).)))).)).)))..)))))))))) ( -39.20, z-score = -6.22, R) >apiMel3.Group16 2947300 86 - 5631066 ---------------CAGUUAAUGCUCUACAUCUACCCUGUAGAUCCGAAUUUGUUUGA---UA---AGAGGCGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUGC- ---------------........((.((((((..(((((.((((.(((((((((((...---..---......))))))))))).)))).)).)))..)))))).))- ( -34.20, z-score = -5.43, R) >triCas2.ChLG2 9946467 80 + 12900155 ----------------------UGCUCUACAUCUACCCUGUAGAUCCGAAUUUGUUUGAAAUCA------GGCGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUGCA ----------------------.((.((((((..(((((.((((.(((((((((((((....))------...))))))))))).)))).)).)))..)))))).)). ( -36.80, z-score = -6.46, R) >consensus _________UU_CUC_UCAGUUUGCUCCACGUCUACCCUGUAGAUCCGAAUUUGUUUUAUACUAG_CUUUAAGGACAAAUUCGGUUCUAGAGAGGUUUGUGUGGUUCA .......................(..((((((..(((((.((((.(((((((((((.................))))))))))).)))).)).)))..))))))..). (-31.12 = -30.72 + -0.40)

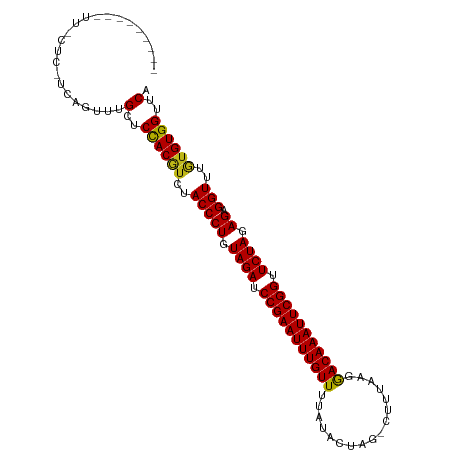
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:23 2011