| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,623,996 – 2,624,086 |
| Length | 90 |
| Max. P | 0.816872 |

| Location | 2,623,996 – 2,624,086 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.31 |
| Shannon entropy | 0.34401 |
| G+C content | 0.47342 |
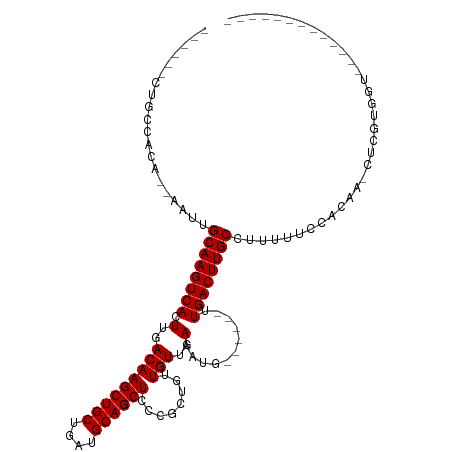
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
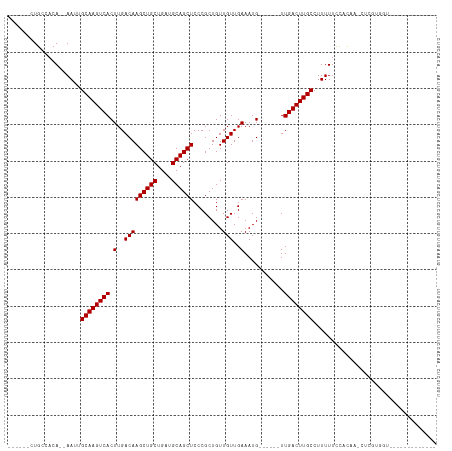
>dm3.chr3R 2623996 90 + 27905053 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCCUUUG-CUCGUUGU------------- ------........--....((((((((((..(((((((((....))))))........)))..)...)------.))))))))............-........------------- ( -21.70, z-score = -0.99, R) >droSim1.chr3R 2654944 90 + 27517382 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCCCCAG-CUCGUUGU------------- ------(((....(--((..((((((((((..(((((((((....))))))........)))..)...)------.))))))))..)))....)))-........------------- ( -23.30, z-score = -1.42, R) >droSec1.super_6 2715057 90 + 4358794 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCCCCAG-CUCGUUGU------------- ------(((....(--((..((((((((((..(((((((((....))))))........)))..)...)------.))))))))..)))....)))-........------------- ( -23.30, z-score = -1.42, R) >droYak2.chr3R 21042948 87 - 28832112 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCCCCAU-CUUGU---------------- ------........--....((((((((((..(((((((((....))))))........)))..)...)------.))))))))............-.....---------------- ( -21.70, z-score = -1.89, R) >droEre2.scaffold_4770 2869741 90 + 17746568 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCCCCAU-CUCGUUGU------------- ------........--....((((((((((..(((((((((....))))))........)))..)...)------.))))))))............-........------------- ( -21.70, z-score = -1.54, R) >droAna3.scaffold_13340 5434658 91 + 23697760 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUUCUUCA--UCGUGCCUU----------- ------........--....((((((((((..(((((((((....))))))........)))..)...)------.))))))))............--.........----------- ( -21.70, z-score = -1.07, R) >dp4.chr2 19759075 96 + 30794189 CUGGCUCUGCCACACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCUACUCUUUUUU---------------- .((((...))))........((((((((((..(((((((((....))))))........)))..)...)------.))))))))..................---------------- ( -24.90, z-score = -1.92, R) >droPer1.super_3 2507288 96 + 7375914 CUGGCUCUGCCACACAAAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUCUACUCUUUUUU---------------- .((((...))))........((((((((((..(((((((((....))))))........)))..)...)------.))))))))..................---------------- ( -24.90, z-score = -1.92, R) >droWil1.scaffold_181089 11990950 91 + 12369635 ------UUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG------UUGACUUGCCUUUUUC----ACCCCGUGUCCCCC--------- ------....((((--((..((((((((((..(((((((((....))))))........)))..)...)------.))))))))..)))..----.....)))......--------- ( -21.81, z-score = -2.04, R) >droVir3.scaffold_13047 6116678 104 + 19223366 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGCUGAUGUUGACUUGCCUUUUUCCAUGAGCUCAUGGCACACACA------ ------.(((((((--(...((..(((....))).((((((....))))))...))..))))......(((.(((..((........))..))).)))....))))......------ ( -29.00, z-score = -0.69, R) >droMoj3.scaffold_6540 16528528 104 + 34148556 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGCUGAUGUUGACUUGCCUUUUUCCAUGAGCUCAUGAAGCACACA------ ------.(((....--....((((((((....(((((((((....))))))...((.(((.....)))))...))))))))))).......((((....))))..)))....------ ( -26.90, z-score = -0.33, R) >droGri2.scaffold_14906 11466007 110 + 14172833 ------CUGCCACA--AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUGCUGAUGUUGACUUGCCUUUUUCCAUGAGCUCAUGAAGCAAGCACACACA ------.(((....--....((((((((....(((((((((....))))))...((.(((.....)))))...))))))))))).......((((....))))..))).......... ( -27.20, z-score = 0.47, R) >consensus ______CUGCCACA__AAUUGCAAGUCACUUGACAAGCUGCUGAUGCAGCUCCCGCUGUUGUUGAAAUG______UUGACUUGCCUUUUUCCACAA_CUCGUGGU_____________ ....................((((((((.......((((((....)))))).(.((....)).)............)))))))).................................. (-20.20 = -20.20 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:21 2011