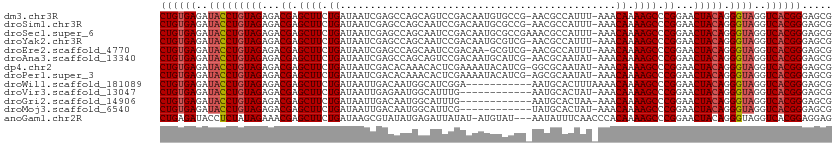
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,602,056 – 2,602,166 |
| Length | 110 |
| Max. P | 0.999690 |

| Location | 2,602,056 – 2,602,166 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.07 |
| Shannon entropy | 0.36049 |
| G+C content | 0.48989 |
| Mean single sequence MFE | -37.31 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.75 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
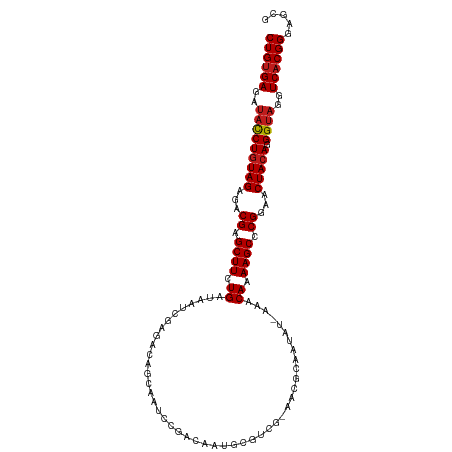
>dm3.chr3R 2602056 110 + 27905053 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AAAUGGCGUU-CGGCACAUUGUCGGACUGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((...-...(((((((-((((.....))))))).))))..........))))))))))...)))))))))..)))).. ( -41.89, z-score = -4.07, R) >droSim1.chr3R 2632932 110 + 27517382 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AAAUGGCGUU-CGGCGCAUUGUCGGAUUGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((..(-((.(((((..-...)))...(((((....))))).)).))).))))))))))...)))))))))..)))).. ( -39.60, z-score = -3.17, R) >droSec1.super_6 2693193 111 + 4358794 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AAAUGGCGUUUCGGCGCAUUGUCGGAUUGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((..(-((.((((((....))))...(((((....))))).)).))).))))))))))...)))))))))..)))).. ( -40.50, z-score = -3.43, R) >droYak2.chr3R 21020629 110 - 28832112 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AAAUGGCGUU-CGACGCAUUGUCGGAUUGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((...-...(((((((-((((.....)))))).)))))..........))))))))))...)))))))))..)))).. ( -39.99, z-score = -3.60, R) >droEre2.scaffold_4770 2845901 109 + 17746568 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AAAUGGCGUU-CGACGC-UUGUCGGAUUGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((...-...(((((((-((((..-..)))))).)))))..........))))))))))...)))))))))..)))).. ( -39.99, z-score = -3.60, R) >droAna3.scaffold_13340 18250907 110 - 23697760 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AUAUUGCGUU-CGAUGCAUUGUCGGACUGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((...-((...(((((-((((.....))))))).)).......))...))))))))))...)))))))))..)))).. ( -39.10, z-score = -3.72, R) >dp4.chr2 11020284 110 - 30794189 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AUAUUGCGCC-CGAUGUAUUUUCGAGUGUUUGUGUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((.((-((((.((((.-(((.......))).))))..)))).))....))))))))))...)))))))))..)))).. ( -37.50, z-score = -4.65, R) >droPer1.super_3 4853605 110 - 7375914 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AUAUUGCGCU-CGAUGUAUUUUCGAGUGUUUGUGUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((.((-((((.(((((-(((.......))))))))..)))).))....))))))))))...)))))))))..)))).. ( -42.30, z-score = -6.10, R) >droWil1.scaffold_181089 11961216 101 + 12369635 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUUUAAAGUGCAUU-----------UCCGAUGCCAUUGUCAAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((......((((((((-----------...)))).)))).........))))))))))...)))))))))..)))).. ( -33.06, z-score = -5.16, R) >droVir3.scaffold_13047 13081408 99 - 19223366 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AUAGUGCAUU------------CAAAUGCCAUUCUCAAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((...-..((.((((.------------...))))....)).......))))))))))...)))))))))..)))).. ( -31.50, z-score = -5.53, R) >droGri2.scaffold_14906 2689383 99 - 14172833 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-UUAGUGCAUU------------CAAAUGCCAUUGUCAAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((...-.((((((((.------------...))).)))))........))))))))))...)))))))))..)))).. ( -33.00, z-score = -5.68, R) >droMoj3.scaffold_6540 17602264 99 - 34148556 CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU-AUAGUGCAUA------------CGAAUGCCAUUGUCAAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((.((-(((((((((.------------...))).)))))).))....))))))))))...)))))))))..)))).. ( -34.40, z-score = -5.16, R) >anoGam1.chr2R 61218901 108 + 62725911 CUCCUCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUGGGUUGAAAUAUU---AUACAU-AUAUAAUCUCAUAUACGCUUAUCAGAAGCUCGUUUCUAUAGAGGUAUCUCAG ........(((..((((((((((...((((((((((((((((((...(((---(((...-.)))))).))).....))))).))))))))))...)))))).))))..))). ( -32.20, z-score = -3.58, R) >consensus CGCUCCCGUGACCUACCCUGUAGUUCCGGGCUUUUGUUU_AUAUUGCGUU_CGACGCAUUGUCGGAUGGCUGGCUCGAUUAUCAGAAGCUCGUCUCUACAGGUAUCUCACAG .......((((..((((.(((((...((((((((((..............................................))))))))))...)))))))))..)))).. (-26.66 = -26.75 + 0.09)
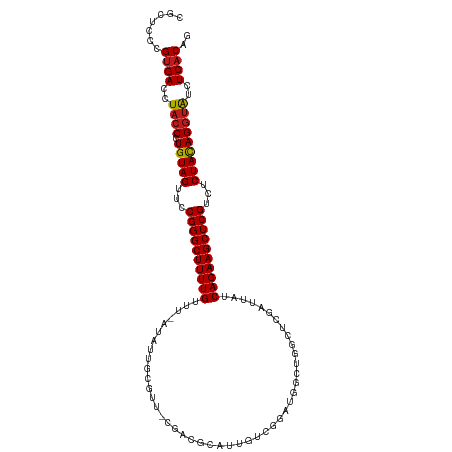
| Location | 2,602,056 – 2,602,166 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.07 |
| Shannon entropy | 0.36049 |
| G+C content | 0.48989 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -22.93 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.962993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2602056 110 - 27905053 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGCCAGCAGUCCGACAAUGUGCCG-AACGCCAUUU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((....(((..(.....)..))).((((.((..-...)))))).-...)).)))).))...))))).))))..))))))..... ( -33.20, z-score = -1.66, R) >droSim1.chr3R 2632932 110 - 27517382 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGCCAGCAAUCCGACAAUGCGCCG-AACGCCAUUU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((....(((...........))).((((.((..-...)))))).-...)).)))).))...))))).))))..))))))..... ( -30.70, z-score = -1.18, R) >droSec1.super_6 2693193 111 - 4358794 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGCCAGCAAUCCGACAAUGCGCCGAAACGCCAUUU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((....(((...........))).((((.((......)))))).-...)).)))).))...))))).))))..))))))..... ( -30.90, z-score = -1.30, R) >droYak2.chr3R 21020629 110 + 28832112 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGCCAGCAAUCCGACAAUGCGUCG-AACGCCAUUU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((......((........))((((.....))))-..........-...)).)))).))...))))).))))..))))))..... ( -31.80, z-score = -1.44, R) >droEre2.scaffold_4770 2845901 109 - 17746568 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGCCAGCAAUCCGACAA-GCGUCG-AACGCCAUUU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((......((........))((((..-..))))-..........-...)).)))).))...))))).))))..))))))..... ( -31.80, z-score = -1.52, R) >droAna3.scaffold_13340 18250907 110 + 23697760 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGCCAGCAGUCCGACAAUGCAUCG-AACGCAAUAU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((....(((..(.....)..)))...(((....-...)))....-...)).)))).))...))))).))))..))))))..... ( -32.70, z-score = -2.07, R) >dp4.chr2 11020284 110 + 30794189 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGACACAAACACUCGAAAAUACAUCG-GGCGCAAUAU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((((......................))))........(((-(((.......-........))))))..))))).))))..))))))..... ( -32.51, z-score = -3.18, R) >droPer1.super_3 4853605 110 + 7375914 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGACACAAACACUCGAAAAUACAUCG-AGCGCAAUAU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((......(.......).(((((.......)))-))........-...)).)))).))...))))).))))..))))))..... ( -30.30, z-score = -2.76, R) >droWil1.scaffold_181089 11961216 101 - 12369635 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUUGACAAUGGCAUCGGA-----------AAUGCACUUUAAAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((....((((.....((((....-----------.))))...))))..)).)))).))...))))).))))..))))))..... ( -29.30, z-score = -2.45, R) >droVir3.scaffold_13047 13081408 99 + 19223366 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUUGAGAAUGGCAUUUG------------AAUGCACUAU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((.......((....((((...------------.)))).))..-...)).)))).))...))))).))))..))))))..... ( -29.40, z-score = -2.95, R) >droGri2.scaffold_14906 2689383 99 + 14172833 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUUGACAAUGGCAUUUG------------AAUGCACUAA-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((.............((((...------------.)))).....-...)).)))).))...))))).))))..))))))..... ( -28.10, z-score = -2.59, R) >droMoj3.scaffold_6540 17602264 99 + 34148556 CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUUGACAAUGGCAUUCG------------UAUGCACUAU-AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((.............((((...------------.)))).....-...)).)))).))...))))).))))..))))))..... ( -28.60, z-score = -2.12, R) >anoGam1.chr2R 61218901 108 - 62725911 CUGAGAUACCUCUAUAGAAACGAGCUUCUGAUAAGCGUAUAUGAGAUUAUAU-AUGUAU---AAUAUUUCAACCCACAAAAGCCCGGAACUACAGGGUAGGUCACGGAGGAG ........(((((..((((......))))(((..(((((((((....)))))-))))..---...................((((.........))))..)))..))))).. ( -21.50, z-score = -0.21, R) >consensus CUGUGAGAUACCUGUAGAGACGAGCUUCUGAUAAUCGAGACAGCAAUCCGACAAUGCGUCG_AACGCAAUAU_AAACAAAAGCCCGGAACUACAGGGUAGGUCACGGGAGCG ((((((..(((((((((...((.((((.((..............................................)).)))).))...))))).))))..))))))..... (-22.93 = -23.17 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:20 2011