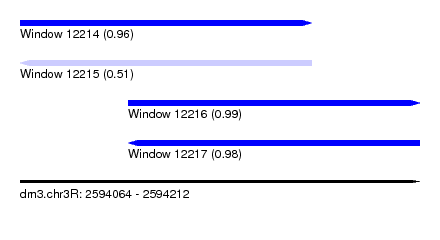
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,594,064 – 2,594,212 |
| Length | 148 |
| Max. P | 0.992641 |

| Location | 2,594,064 – 2,594,172 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.67 |
| Shannon entropy | 0.67336 |
| G+C content | 0.37753 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -8.07 |
| Energy contribution | -7.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
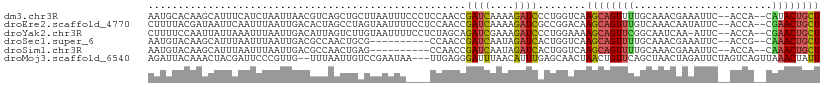
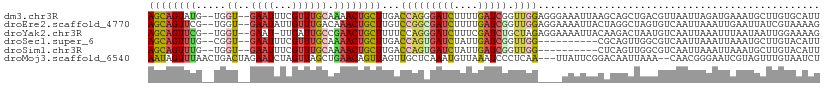
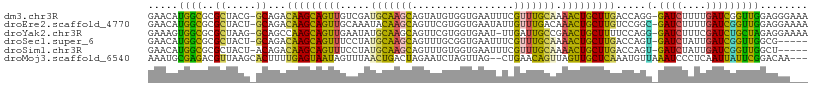
>dm3.chr3R 2594064 108 + 27905053 AAUGCACAAGCAUUUCAUCUAAUUAACGUCAGCUGCUUAAUUUCCCUCCAACCGAUCAAAAGAUCCCUGGUCAAGCAGUUUUGCAAACGAAAUUC--ACCA--CAUACUGCU (((((....))))).............((.((((((((.........(((...((((....))))..)))..))))))))..))...........--....--......... ( -16.10, z-score = -0.82, R) >droEre2.scaffold_4770 2836932 108 + 17746568 CUUUUACGAUAAUUCAAUUUAAUUGACACUAGCCUAGUAAUUUUCCUCCAACCGAUCAAAAGAUCGCCGGACAAGCAGUUUGUCAAACAAUAUUC--ACCA--CGAACUGCU .............(((((...)))))....................(((...(((((....)))))..)))..((((((((((............--...)--))))))))) ( -18.86, z-score = -2.40, R) >droYak2.chr3R 21011859 107 - 28832112 CUUUUCCAAUUAUUAAAUUUAAUUGACAUUAGUCUUGUAAUUUUCCUCUAGCAGAUCGAAAGAUCCCUGGAAAAGCAGUUCGGCAAUCAA-AUUC--ACCA--CGAACUGCU .(((((((......(((.(((...(((....)))...))).))).........((((....))))..)))))))((((((((........-....--....--)))))))). ( -27.69, z-score = -4.23, R) >droSec1.super_6 2685063 98 + 4358794 AAUGUACAAGCAUUUAAUUUAAUUGACGCCAACUGCG----------CCAACCGAUCAAUAGAUCACUGGUCAAGCAGUUUUGCAAACGAAAUUC--ACCG--CAAACUGCU (((((....)))))........(((((((.....)).----------......((((....))))....)))))(((((((.((...........--...)--)))))))). ( -20.14, z-score = -1.25, R) >droSim1.chr3R 2624844 98 + 27517382 AAUGUACAAGCAUUUAAUUUAAUUGACGCCAACUGAG----------CCAACCGAUCAAUAGAUCACUGGUCAAGCAGUUUUGCAAACGAAAUUC--ACCA--CAAACUGCU (((((....)))))........((((((((....).)----------).....((((....))))....)))))(((((((((............--..))--.))))))). ( -16.44, z-score = -0.67, R) >droMoj3.scaffold_6540 17591712 107 - 34148556 AGAUUACAAACUACGAUUCCCGUUG--UUUAAUUGUCCGAAUAA---UUGAGGGAUUUAACAUUUGAGCAACUAACUGUUCAGCUAACUAGAUUCUAGUCAGUUAAACUAUU .(((...((((.(((.....))).)--)))....)))....(((---((((((((((((....(((((((......))))))).....))))))))..)))))))....... ( -20.60, z-score = -1.20, R) >consensus AAUGUACAAGCAUUUAAUUUAAUUGACGCCAACUGAGUAAUUU____CCAACCGAUCAAAAGAUCACUGGACAAGCAGUUUUGCAAACGAAAUUC__ACCA__CAAACUGCU .....................................................((((....))))........((((((((.......................)))))))) ( -8.07 = -7.93 + -0.14)
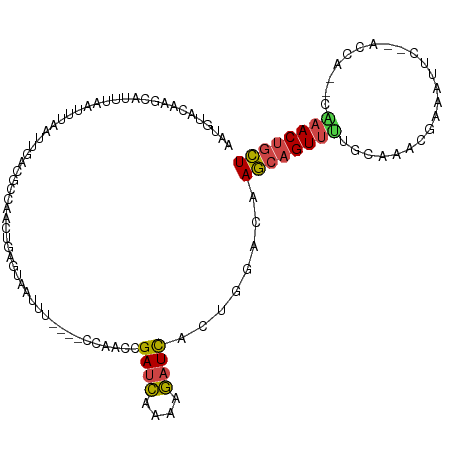
| Location | 2,594,064 – 2,594,172 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 63.67 |
| Shannon entropy | 0.67336 |
| G+C content | 0.37753 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -9.37 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2594064 108 - 27905053 AGCAGUAUG--UGGU--GAAUUUCGUUUGCAAAACUGCUUGACCAGGGAUCUUUUGAUCGGUUGGAGGGAAAUUAAGCAGCUGACGUUAAUUAGAUGAAAUGCUUGUGCAUU ..((((.((--(...--.((((((.((((((....)))..((((...((((....)))))))).))).))))))..))))))).((((.....)))).(((((....))))) ( -24.50, z-score = 0.20, R) >droEre2.scaffold_4770 2836932 108 - 17746568 AGCAGUUCG--UGGU--GAAUAUUGUUUGACAAACUGCUUGUCCGGCGAUCUUUUGAUCGGUUGGAGGAAAAUUACUAGGCUAGUGUCAAUUAAAUUGAAUUAUCGUAAAAG .....((..--((((--((..(((..((((((....(((((((((((((((....)))).))))))(........))))))...))))))...)))....))))))..)).. ( -25.00, z-score = -1.02, R) >droYak2.chr3R 21011859 107 + 28832112 AGCAGUUCG--UGGU--GAAU-UUGAUUGCCGAACUGCUUUUCCAGGGAUCUUUCGAUCUGCUAGAGGAAAAUUACAAGACUAAUGUCAAUUAAAUUUAAUAAUUGGAAAAG .(((((((.--.((.--.(..-....)..)))))))))(((((((((((((....)))))..................(((....)))...............)))))))). ( -28.60, z-score = -2.58, R) >droSec1.super_6 2685063 98 - 4358794 AGCAGUUUG--CGGU--GAAUUUCGUUUGCAAAACUGCUUGACCAGUGAUCUAUUGAUCGGUUGG----------CGCAGUUGGCGUCAAUUAAAUUAAAUGCUUGUACAUU ((((((((.--..(.--.(((...)))..).))))))))((((.(((((((....))))((((((----------(((.....))))))))).........))).)).)).. ( -29.60, z-score = -1.96, R) >droSim1.chr3R 2624844 98 - 27517382 AGCAGUUUG--UGGU--GAAUUUCGUUUGCAAAACUGCUUGACCAGUGAUCUAUUGAUCGGUUGG----------CUCAGUUGGCGUCAAUUAAAUUAAAUGCUUGUACAUU ((((((((.--..(.--.(((...)))..).))))))))((((((.(((((....))))).).))----------.)))((.(((((............)))))...))... ( -24.40, z-score = -0.91, R) >droMoj3.scaffold_6540 17591712 107 + 34148556 AAUAGUUUAACUGACUAGAAUCUAGUUAGCUGAACAGUUAGUUGCUCAAAUGUUAAAUCCCUCAA---UUAUUCGGACAAUUAAA--CAACGGGAAUCGUAGUUUGUAAUCU ....(((((.((((((((...)))))))).)))))..(((((((..(.((((.............---.)))).)..)))))))(--(.(((.....))).))......... ( -19.14, z-score = -0.63, R) >consensus AGCAGUUUG__UGGU__GAAUUUCGUUUGCAAAACUGCUUGACCAGCGAUCUUUUGAUCGGUUGG____AAAUUACACAGCUAACGUCAAUUAAAUUAAAUGCUUGUAAAUU ((((((((.......................))))))))...(((((((((....))))).))))............................................... ( -9.37 = -9.52 + 0.15)

| Location | 2,594,104 – 2,594,212 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.61173 |
| G+C content | 0.45249 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -15.38 |
| Energy contribution | -14.78 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
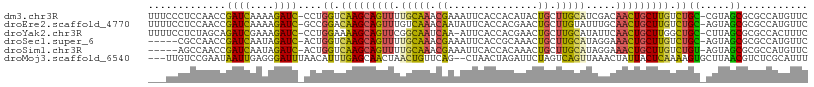
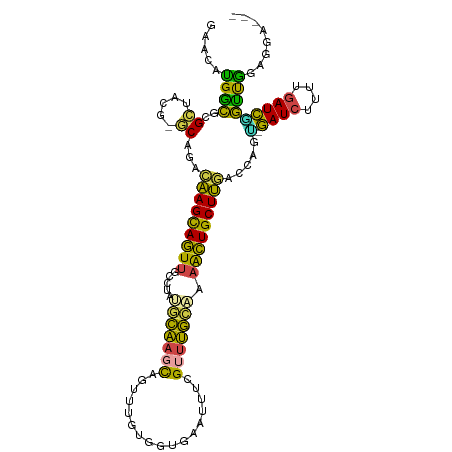
>dm3.chr3R 2594104 108 + 27905053 UUUCCCUCCAACCGAUCAAAAGAUC-CCUGGUCAAGCAGUUUUGCAAACGAAAUUCACCACAUACUGCUUGCAUCGACAACUGCUUGUCUGC-CGUAGCGCGCCAUGUUC .............((((....))))-..(((((((((((((.(((((.((...............)).))))).....)))))))))...((-......))))))..... ( -21.06, z-score = -0.43, R) >droEre2.scaffold_4770 2836972 108 + 17746568 UUUUCCUCCAACCGAUCAAAAGAUC-GCCGGACAAGCAGUUUGUCAAACAAUAUUCACCACGAACUGCUUGUAUUUGCAACUGCUUGUCUGC-AGUAGCGCGCCAUGUUC ............(((((....))))-)..(((((((((((((((...............)))))))))))))....((.(((((......))-))).))...))...... ( -33.96, z-score = -4.07, R) >droYak2.chr3R 21011899 107 - 28832112 UUUUCCUCUAGCAGAUCGAAAGAUC-CCUGGAAAAGCAGUUCGGCAAUCAA-AUUCACCACGAACUGCUUGCAUAUUCAACUGCUUGGCUGC-CUUAGCGCGCCACUUUC ......(((((..((((....))))-.))))).((((((((((........-........))))))))))(((........))).(((((((-....))).))))..... ( -34.59, z-score = -3.71, R) >droSec1.super_6 2685098 103 + 4358794 -----CGCCAACCGAUCAAUAGAUC-ACUGGUCAAGCAGUUUUGCAAACGAAAUUCACCGCAAACUGCUUGCAUAGGAAACUGCUUGUCUGC-AGUAGCGCGCCAUGUUC -----((((..((((((....))))-....(.((((((((((.((..............)))))))))))))...))..(((((......))-))).).)))........ ( -30.24, z-score = -1.64, R) >droSim1.chr3R 2624879 103 + 27517382 -----AGCCAACCGAUCAAUAGAUC-ACUGGUCAAGCAGUUUUGCAAACGAAAUUCACCACAAACUGCUUGCAUAGGAAACUGCUUGUCUGU-AGUAGCGCGCCAUGUUC -----.(((....((((....))))-(((((.(((((((((((((((.((...............)).)))))....)))))))))).))..-))).).))......... ( -24.16, z-score = -0.30, R) >droMoj3.scaffold_6540 17591742 105 - 34148556 ---UUGUCCGAAUAAUUGAGGGAUUUAACAUUUGAGCAACUAACUGUUCAG--CUAACUAGAUUCUAGUCAGUUAAACUAUUACUCAAAAGUGCUUAACGUCUCGCAUUU ---.............((.(((((((((((((((((((......)))))((--((.(((((...))))).))))..............))))).)))).))))).))... ( -19.50, z-score = -0.64, R) >consensus ___UCCUCCAACCGAUCAAAAGAUC_ACUGGUCAAGCAGUUUUGCAAACAAAAUUCACCACAAACUGCUUGCAUAGGCAACUGCUUGUCUGC_AGUAGCGCGCCAUGUUC ......((((...((((....))))...))))(((((((((.(((((.((...............)).))))).....)))))))))....................... (-15.38 = -14.78 + -0.60)

| Location | 2,594,104 – 2,594,212 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.61173 |
| G+C content | 0.45249 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -20.17 |
| Energy contribution | -18.30 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2594104 108 - 27905053 GAACAUGGCGCGCUACG-GCAGACAAGCAGUUGUCGAUGCAAGCAGUAUGUGGUGAAUUUCGUUUGCAAAACUGCUUGACCAGG-GAUCUUUUGAUCGGUUGGAGGGAAA .(((.(((...((....-))...(((((((((.....(((((((...((.......))...))))))).))))))))).)))..-((((....)))).)))......... ( -30.50, z-score = -0.13, R) >droEre2.scaffold_4770 2836972 108 - 17746568 GAACAUGGCGCGCUACU-GCAGACAAGCAGUUGCAAAUACAAGCAGUUCGUGGUGAAUAUUGUUUGACAAACUGCUUGUCCGGC-GAUCUUUUGAUCGGUUGGAGGAAAA ...........((.(((-((......))))).)).....(((((((((....((.((......)).)).)))))))))((((((-((((....)))).))))))...... ( -34.90, z-score = -1.88, R) >droYak2.chr3R 21011899 107 + 28832112 GAAAGUGGCGCGCUAAG-GCAGCCAAGCAGUUGAAUAUGCAAGCAGUUCGUGGUGAAU-UUGAUUGCCGAACUGCUUUUCCAGG-GAUCUUUCGAUCUGCUAGAGGAAAA .....((((..((....-)).))))((((((((((.....(((((((((..((..(..-....)..))))))))))).((....-))...))))).)))))......... ( -33.90, z-score = -0.93, R) >droSec1.super_6 2685098 103 - 4358794 GAACAUGGCGCGCUACU-GCAGACAAGCAGUUUCCUAUGCAAGCAGUUUGCGGUGAAUUUCGUUUGCAAAACUGCUUGACCAGU-GAUCUAUUGAUCGGUUGGCG----- ...(((((.(....(((-((......)))))..))))))((((((((((...(..(((...)))..).)))))))))).(((.(-((((....))))).).))..----- ( -33.00, z-score = -1.13, R) >droSim1.chr3R 2624879 103 - 27517382 GAACAUGGCGCGCUACU-ACAGACAAGCAGUUUCCUAUGCAAGCAGUUUGUGGUGAAUUUCGUUUGCAAAACUGCUUGACCAGU-GAUCUAUUGAUCGGUUGGCU----- ...........((((((-.....((((((((((....((((((((((((.....)))))..)))))))))))))))))......-((((....)))))).)))).----- ( -29.70, z-score = -0.76, R) >droMoj3.scaffold_6540 17591742 105 + 34148556 AAAUGCGAGACGUUAAGCACUUUUGAGUAAUAGUUUAACUGACUAGAAUCUAGUUAG--CUGAACAGUUAGUUGCUCAAAUGUUAAAUCCCUCAAUUAUUCGGACAA--- ......(((..(((.((((..((((((((((.(((((.((((((((...))))))))--.))))).....)))))))))))))).)))..)))..............--- ( -26.90, z-score = -2.73, R) >consensus GAACAUGGCGCGCUACG_GCAGACAAGCAGUUGCCUAUGCAAGCAGUUUGUGGUGAAUUUCGUUUGCAAAACUGCUUGACCAGU_GAUCUUUUGAUCGGUUGGAGGA___ .....((((..............(((((((((.....(((((((.................))))))).))))))))).......((((....)))).))))........ (-20.17 = -18.30 + -1.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:18 2011