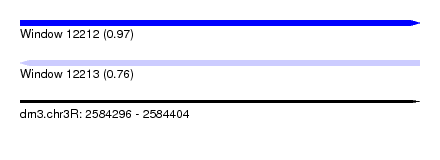
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,584,296 – 2,584,404 |
| Length | 108 |
| Max. P | 0.968233 |

| Location | 2,584,296 – 2,584,404 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.57118 |
| G+C content | 0.36795 |
| Mean single sequence MFE | -18.75 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
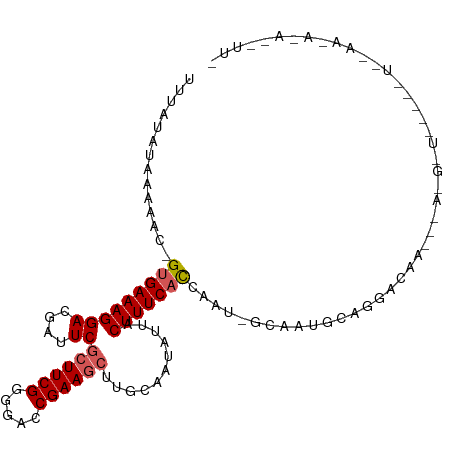
>dm3.chr3R 2584296 108 + 27905053 UUUAUAUAAAAAC-GUGAAAGGACGAUUCGCUUCGGGGACCGAAUCUUGCAAUAUUACUUUCACUAAUCAUCAUGCAGGACAACA-AUGCUUGUUUUAGAAAAUAUGUUG ..(((((......-(((((((....(((.(((((((...)))))....)))))....)))))))...((.....(((((.((...-.)))))))....))..)))))... ( -18.70, z-score = -0.45, R) >droSim1.chr3R 2614983 81 + 27517382 UUCAUAUAAAAAC-GUGAAAGGACGAUUCGCUUCGGGGACCGAAGCUUGCAAUAUUACUUUCACCAAU-GCAAUGCAGGAUAA--------------------------- (((..........-(((((((((....))(((((((...)))))))...........)))))))...(-((...))))))...--------------------------- ( -18.50, z-score = -1.69, R) >droSec1.super_6 2675512 81 + 4358794 UUCAUAUAAAAAC-GUGAAAGGACGAUUCGCUUCGGGGACCGAAGCUUGCAAUAUUACUUUCACCAAU-GCAAUGCAGGAUAA--------------------------- (((..........-(((((((((....))(((((((...)))))))...........)))))))...(-((...))))))...--------------------------- ( -18.50, z-score = -1.69, R) >droYak2.chr3R 21001689 110 - 28832112 UUUAUAUAAAAAAAGUGAAAGGACGAUUCGCUUCGUGGACCGAAGCUUGCAAUAUCCCUUUCAUCAGCAUCUCAAAAAUACAUUGCAGGAUAACAAUCAAAUAUAUAUUA ..((((((......((((((((...(((.((((((.....))))))....)))...))))))))..(((..............)))...............))))))... ( -20.04, z-score = -1.59, R) >droEre2.scaffold_4770 2826795 107 + 17746568 UUUAUAUAAAAAA-GUGAAAGGACG-UUCACUUCGAAGACCGAAGCUUGCAAUAUCCCUUUCCUCAGCUUUUCAGAAAUGCAA-GCAGGAUAACAAUUGAAUAAAACUUA ...........((-((....(((..-(((((((((.....)))))..)).))..)))...((((..((((..((....)).))-))))))...............)))). ( -18.00, z-score = -0.61, R) >consensus UUUAUAUAAAAAC_GUGAAAGGACGAUUCGCUUCGGGGACCGAAGCUUGCAAUAUUACUUUCACCAAU_GCAAUGCAGGACAA___A_G_U_____U__AA_A_A__UU_ ..............(((((((((....))((((((.....))))))...........))))))).............................................. (-12.66 = -13.02 + 0.36)

| Location | 2,584,296 – 2,584,404 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.57118 |
| G+C content | 0.36795 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -11.62 |
| Energy contribution | -12.26 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
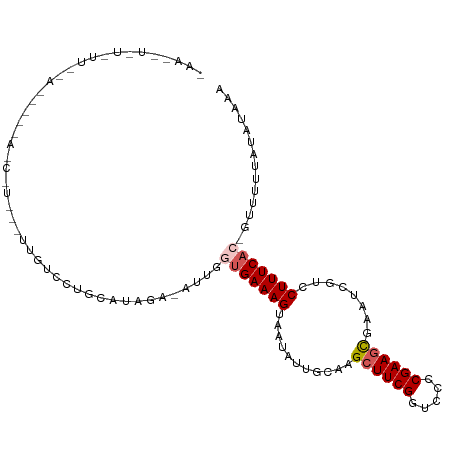
>dm3.chr3R 2584296 108 - 27905053 CAACAUAUUUUCUAAAACAAGCAU-UGUUGUCCUGCAUGAUGAUUAGUGAAAGUAAUAUUGCAAGAUUCGGUCCCCGAAGCGAAUCGUCCUUUCAC-GUUUUUAUAUAAA ....((((.....(((((...(((-..(.(.....))..)))....(((((((.....((((....(((((...)))))))))......)))))))-))))).))))... ( -18.20, z-score = -0.48, R) >droSim1.chr3R 2614983 81 - 27517382 ---------------------------UUAUCCUGCAUUGC-AUUGGUGAAAGUAAUAUUGCAAGCUUCGGUCCCCGAAGCGAAUCGUCCUUUCAC-GUUUUUAUAUGAA ---------------------------..............-....(((((((...........(((((((...)))))))........)))))))-............. ( -17.51, z-score = -1.14, R) >droSec1.super_6 2675512 81 - 4358794 ---------------------------UUAUCCUGCAUUGC-AUUGGUGAAAGUAAUAUUGCAAGCUUCGGUCCCCGAAGCGAAUCGUCCUUUCAC-GUUUUUAUAUGAA ---------------------------..............-....(((((((...........(((((((...)))))))........)))))))-............. ( -17.51, z-score = -1.14, R) >droYak2.chr3R 21001689 110 + 28832112 UAAUAUAUAUUUGAUUGUUAUCCUGCAAUGUAUUUUUGAGAUGCUGAUGAAAGGGAUAUUGCAAGCUUCGGUCCACGAAGCGAAUCGUCCUUUCACUUUUUUUAUAUAAA ...((((((....(((((......)))))(((((.....)))))...((((((((.........((((((.....))))))......)))))))).......)))))).. ( -25.26, z-score = -1.79, R) >droEre2.scaffold_4770 2826795 107 - 17746568 UAAGUUUUAUUCAAUUGUUAUCCUGC-UUGCAUUUCUGAAAAGCUGAGGAAAGGGAUAUUGCAAGCUUCGGUCUUCGAAGUGAA-CGUCCUUUCAC-UUUUUUAUAUAAA ..((((((..(((..(((........-..)))....)))))))))(((..((((((..((....((((((.....)))))).))-..))))))..)-))........... ( -19.30, z-score = 0.35, R) >consensus _AA__U_U_UU__A_____A_C_U___UUGUCCUGCAUAGA_AUUGGUGAAAGUAAUAUUGCAAGCUUCGGUCCCCGAAGCGAAUCGUCCUUUCAC_GUUUUUAUAUAAA ..............................................(((((((...........((((((.....))))))((....))))))))).............. (-11.62 = -12.26 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:15 2011