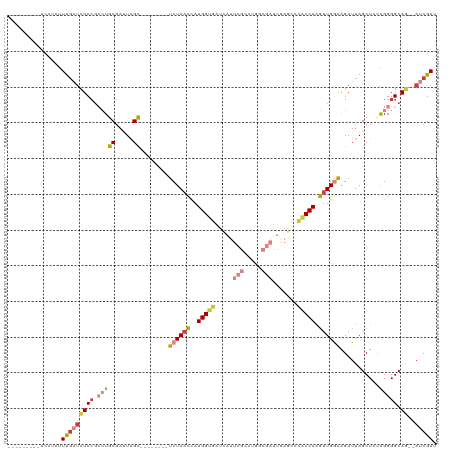
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 632,152 – 632,257 |
| Length | 105 |
| Max. P | 0.810357 |

| Location | 632,152 – 632,253 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.93 |
| Shannon entropy | 0.53797 |
| G+C content | 0.50975 |
| Mean single sequence MFE | -36.21 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.06 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 632152 101 + 23011544 ---------UCCUCUUUGAUCUCCGCCUGCGAUUAGC--------UCUAACUAUGGACAUCAUUCGCCUGGCGAUUAAGCCCACCAAGUUGGAAGAUUAGCUGCAGGGGCAG--AUCGAU ---------......(((((((((.((((((.(((((--------(((((((.(((.......((((...)))).........))))))))).)).)))).)))))))).))--))))). ( -34.19, z-score = -1.95, R) >droEre2.scaffold_4929 690169 85 + 26641161 ---------UCCUCUUUGAUCUCCGCCUGCGAUUAGC--------UCUAACCAUGGUGAUUA----------------GCCCACCAAGUUGGAGGAUUAGCUUUAGGGGCAG--AUCGAU ---------......(((((((((.(((((((((..(--------((((((..(((((....----------------...))))).))))))))))).))...))))).))--))))). ( -28.60, z-score = -1.82, R) >droYak2.chr2L 622134 85 + 22324452 ---------UCCUCUUUGAUCUCCGCCUGCGAUUAGC--------UCUAACCAUGGUGAUCA----------------GCCCACCAAGAUGGAAGAUUAGCUCUAGGGGCAG--AUCGAU ---------......(((((((((.((((.((((((.--------(((..((((((((....----------------...)))))...))).))))))).)))))))).))--))))). ( -25.00, z-score = -0.63, R) >droSec1.super_14 614274 96 + 2068291 --------------UUUGAUCUCCUCCUGCGAUUAGC--------UCUAACCAUGGUGAUCAUUCGCCAGGCGAUUGAGCCCACCAAGUUGAAAGAUUAGCUGCAGGGGCAG--AUCGAU --------------.(((((((.(((((((.....((--------((...((.((((((....)))))))).....))))......((((((....))))))))))))).))--))))). ( -39.30, z-score = -3.97, R) >droSim1.chr2L 638643 101 + 22036055 ---------UCCUCUUUGAUCUCCGCCUGCGAUUAGC--------UCUAACCAUGGUGAUCAUUCGCCAGGCGAUUGAACCCACCAAGUUGAAAGAUUAGCUGCAGGGGCAG--AUCGAU ---------......(((((((((.((((((.(((((--------(.((((..(((((.(((.((((...)))).)))...))))).))))..)).)))).)))))))).))--))))). ( -35.80, z-score = -2.74, R) >dp4.chr4_group3 2595647 120 - 11692001 GUCUCCGUCUCCAUCUCGGAGUCCGAGUCCGAGUCGGAGUCGGAGUCUAAUCAUGGUGAUCAUUCGAUUUGCGAUUGAGCGUGCCACAUUGGGAGAUUAGCUCUGGGGGCAUUCAACGAU (((((((.((((..((((((.......))))))..)))).)((((.((((((.(((((.(((.(((.....))).)))...)))))(....)..)))))))))))))))).......... ( -46.30, z-score = -1.65, R) >droPer1.super_8 3685257 120 - 3966273 GUCUCCGUCUCCAUCUCCAUCUCGGAGUCCGAGUCGGAGUCGGAGUCUAAUCAUGGUGAUCAUUCGAUUUGCGAUUGAGCGUGCCACAUUGGGAGAUUAGCUCUGGGGGCAUUCAACGAU (((((((.((((..((((..(((((...)))))..))))..)))).)......(((((.(((.(((.....))).)))...))))).....((((.....)))))))))).......... ( -44.30, z-score = -1.38, R) >consensus _________UCCUCUUUGAUCUCCGCCUGCGAUUAGC________UCUAACCAUGGUGAUCAUUCGCCUGGCGAUUGAGCCCACCAAGUUGGAAGAUUAGCUCUAGGGGCAG__AUCGAU ................((((((......((.....))........((((((..(((((.....(((.....))).......))))).))))))))))))(((.....))).......... (-12.00 = -12.06 + 0.06)


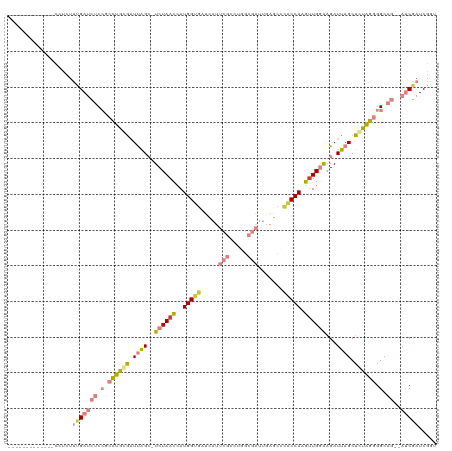
| Location | 632,153 – 632,257 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.38 |
| Shannon entropy | 0.57453 |
| G+C content | 0.52431 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.16 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 632153 104 + 23011544 -------------CCUCUUUGAUCUCCGCCUGCGAUUAGC-UCUAACUAUGGACAUCAUUCGCCUGGCGAUUAAGCCCACCAAGUUGGAAGAUUAGCUGCAGGGGCAG--AUCGAUCGGU -------------((...(((((((((.((((((.(((((-(((((((.(((.......((((...)))).........))))))))).)).)))).)))))))).))--)))))..)). ( -35.49, z-score = -1.68, R) >droEre2.scaffold_4929 690170 88 + 26641161 -------------CCUCUUUGAUCUCCGCCUGCGAUUAGC-UCUAACCAUGGUGAUUA----------------GCCCACCAAGUUGGAGGAUUAGCUUUAGGGGCAG--AUCGAUCGGU -------------((...(((((((((.(((((((((..(-((((((..(((((....----------------...))))).))))))))))).))...))))).))--)))))..)). ( -29.90, z-score = -1.54, R) >droYak2.chr2L 622135 88 + 22324452 -------------CCUCUUUGAUCUCCGCCUGCGAUUAGC-UCUAACCAUGGUGAUCA----------------GCCCACCAAGAUGGAAGAUUAGCUCUAGGGGCAG--AUCGAUCGGG -------------(((..(((((((((.((((.((((((.-(((..((((((((....----------------...)))))...))).))))))).)))))))).))--)))))..))) ( -28.40, z-score = -0.87, R) >droSec1.super_14 614274 100 + 2068291 -----------------UUUGAUCUCCUCCUGCGAUUAGC-UCUAACCAUGGUGAUCAUUCGCCAGGCGAUUGAGCCCACCAAGUUGAAAGAUUAGCUGCAGGGGCAG--AUCGAUCGGU -----------------.(((((((.(((((((.....((-((...((.((((((....)))))))).....))))......((((((....))))))))))))).))--)))))..... ( -39.30, z-score = -3.13, R) >droSim1.chr2L 638644 104 + 22036055 -------------CCUCUUUGAUCUCCGCCUGCGAUUAGC-UCUAACCAUGGUGAUCAUUCGCCAGGCGAUUGAACCCACCAAGUUGAAAGAUUAGCUGCAGGGGCAG--AUCGAUCGGU -------------((...(((((((((.((((((.(((((-(.((((..(((((.(((.((((...)))).)))...))))).))))..)).)))).)))))))).))--)))))..)). ( -37.10, z-score = -2.39, R) >dp4.chr4_group3 2595651 118 - 11692001 CCGUCUCCAUCUCGGAGUCCGAGUCCGAGUCGGAGUCGGAGUCUAAUCAUGGUGAUCAUUCGAUUUGCGAUUGAGCGUGCCACAUUGGGAGAUUAGCUCUGGGGGCAUUCAACGAUCG-- ..((((((((((((((.(((((.((((...)))).))))).))).....(((((.(((.(((.....))).)))...)))))......)))).......)))))))............-- ( -41.91, z-score = -0.63, R) >droPer1.super_8 3685261 118 - 3966273 CCGUCUCCAUCUCCAUCUCGGAGUCCGAGUCGGAGUCGGAGUCUAAUCAUGGUGAUCAUUCGAUUUGCGAUUGAGCGUGCCACAUUGGGAGAUUAGCUCUGGGGGCAUUCAACGAUCG-- ..((((((..((((..(((((...)))))..))))..((((.((((((.(((((.(((.(((.....))).)))...)))))(....)..))))))))))))))))............-- ( -42.20, z-score = -1.00, R) >consensus _____________CCUCUUUGAUCUCCGCCUGCGAUUAGC_UCUAACCAUGGUGAUCAUUCGCCUGGCGAUUGAGCCCACCAAGUUGGAAGAUUAGCUCUAGGGGCAG__AUCGAUCGGU ....................((((.((.((((((.((((..((((((..(((((.....(((.....))).......))))).))))))...)))).))))))))........))))... (-15.44 = -15.16 + -0.29)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:29 2011