
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,534,705 – 2,534,811 |
| Length | 106 |
| Max. P | 0.579771 |

| Location | 2,534,705 – 2,534,811 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 62.85 |
| Shannon entropy | 0.69226 |
| G+C content | 0.46647 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -12.84 |
| Energy contribution | -14.40 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
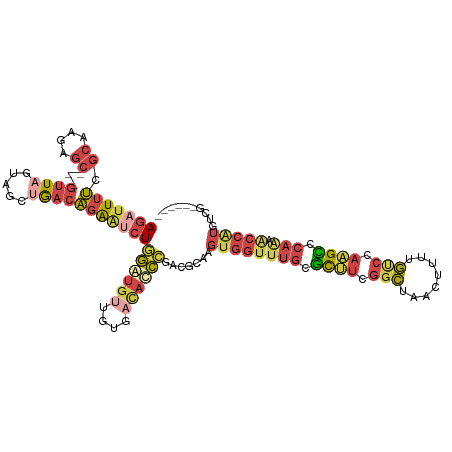
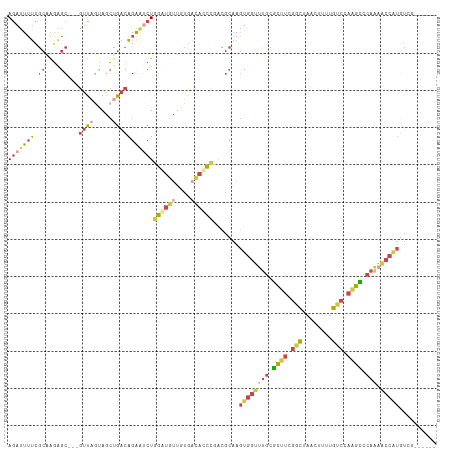
>dm3.chr3R 2534705 106 + 27905053 AGAUUAUCGCAGGAGC---AUUAAUAGCUGACAGAAUCUGGGUGUUGUGACACCCUAAGCAAGUGGUUUGCUCUUCGGCUAACUUUUGUCGAAGACCAAAACCGUGUAU------ (((((....(((....---........)))....)))))((((((....))))))...(((...(((((..((((((((........))))))))...))))).)))..------ ( -32.10, z-score = -1.97, R) >droWil1.scaffold_181089 11875300 91 + 12369635 AAACUUAUCGAAUGGC--AGAUGAGAGA-AACUGGA-CUAAAAACUCUAACAUUUUGGAAGAAUUGAAUGGAUAAUUAUUAACAAUAAAUCGUGU-------------------- ...((((((.......--.)))))).((-...((((-........)))).(((((..........)))))...................))....-------------------- ( -8.40, z-score = 0.81, R) >droEre2.scaffold_4770 2776787 106 + 17746568 AGAUUUUCGCGAGGGC--GGUU-GCAGCUGACAGGAGCUAGGUGUUAUGUCACCCGGCGCAAGUGGUUUGUGCUUCGGCUAACGCAUGCCGAAGCUCAGAACCAUGUCG------ ......((((....))--))..-......(((....((..((((......))))..))....(((((((..((((((((........))))))))...)))))))))).------ ( -42.30, z-score = -1.83, R) >droYak2.chr3R 20951813 115 - 28832112 AGAUUUUUGCAAGGGCAAGGUUAGUAGCUGACAGGGUCUGGGUGUUGUGACACCCCAUGCAAGUGGUUUGUGCCUCGGCUAACGCAUGUCGAAGCCCAGCACCAUAACCGUAUCG .(((..(((((.((((...(((((...)))))...))))((((((....))))))..)))))((((((((.((.(((((........))))).)).))).)))))......))). ( -38.80, z-score = -0.74, R) >droSec1.super_6 2627871 106 + 4358794 AGAUUUUCGCAAGAGC---GUUAGUAACUGACAGAGUCUGGAUGUUGUGAUACCCGACGCAAGUGGUUUGCGCUUCGGCUAAAUUUUGUCCAAGCCCAAAGCCAUGUGG------ ......((....))((---(((((...)))))...(((.((.(((....))))).)))))..(((((((..((((.(((........))).))))...)))))))....------ ( -28.30, z-score = -0.42, R) >droSim1.chr3R 2567062 106 + 27517382 AGAUUUUCGCAAGAGC---GUUAGUAACUGACAGAGUCUGGAUGUUGUGAUACCCGACGCAAGUGGUUUGCGCUUCGGCUAACUUUUGUCCAAGCCCAAAACCAUGUGG------ ......((....))((---(((((...)))))...(((.((.(((....))))).)))))..(((((((..((((.(((........))).))))...)))))))....------ ( -28.30, z-score = -0.43, R) >consensus AGAUUUUCGCAAGAGC___GUUAGUAGCUGACAGAAUCUGGAUGUUGUGACACCCGACGCAAGUGGUUUGCGCUUCGGCUAACUUUUGUCCAAGCCCAAAACCAUGUCG______ ........((.........((((.....)))).......((((((....))))))...))..(((((((..((((.(((........))).))))...))))))).......... (-12.84 = -14.40 + 1.56)

| Location | 2,534,705 – 2,534,811 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 62.85 |
| Shannon entropy | 0.69226 |
| G+C content | 0.46647 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -9.61 |
| Energy contribution | -9.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
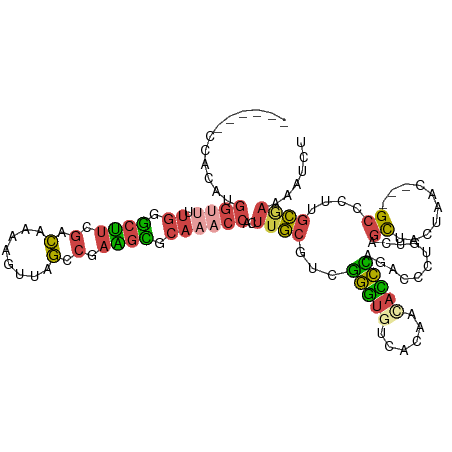
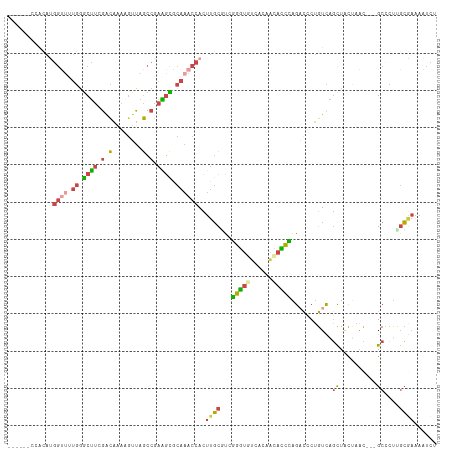
>dm3.chr3R 2534705 106 - 27905053 ------AUACACGGUUUUGGUCUUCGACAAAAGUUAGCCGAAGAGCAAACCACUUGCUUAGGGUGUCACAACACCCAGAUUCUGUCAGCUAUUAAU---GCUCCUGCGAUAAUCU ------......(((((.(.((((((.(........).)))))).))))))..((((...((((((....))))))..........(((.......---)))...))))...... ( -28.30, z-score = -1.91, R) >droWil1.scaffold_181089 11875300 91 - 12369635 --------------------ACACGAUUUAUUGUUAAUAAUUAUCCAUUCAAUUCUUCCAAAAUGUUAGAGUUUUUAG-UCCAGUU-UCUCUCAUCU--GCCAUUCGAUAAGUUU --------------------....(((((((((.................((((((..(.....)..)))))).....-..(((..-........))--).....))))))))). ( -8.70, z-score = -0.66, R) >droEre2.scaffold_4770 2776787 106 - 17746568 ------CGACAUGGUUCUGAGCUUCGGCAUGCGUUAGCCGAAGCACAAACCACUUGCGCCGGGUGACAUAACACCUAGCUCCUGUCAGCUGC-AACC--GCCCUCGCGAAAAUCU ------.(((((((((.((.((((((((........)))))))).))))))).....((..((((......))))..))...))))......-...(--((....)))....... ( -37.90, z-score = -2.78, R) >droYak2.chr3R 20951813 115 + 28832112 CGAUACGGUUAUGGUGCUGGGCUUCGACAUGCGUUAGCCGAGGCACAAACCACUUGCAUGGGGUGUCACAACACCCAGACCCUGUCAGCUACUAACCUUGCCCUUGCAAAAAUCU .(((..((((...(((((.((((.((.....))..))))..))))).))))..(((((.(((((((....)))))).(((...)))................).)))))..))). ( -35.10, z-score = -0.78, R) >droSec1.super_6 2627871 106 - 4358794 ------CCACAUGGCUUUGGGCUUGGACAAAAUUUAGCCGAAGCGCAAACCACUUGCGUCGGGUAUCACAACAUCCAGACUCUGUCAGUUACUAAC---GCUCUUGCGAAAAUCU ------.....((((...(((..((((.......((.(((..((((((.....))))))))).))........))))..))).))))........(---((....)))....... ( -26.96, z-score = -1.19, R) >droSim1.chr3R 2567062 106 - 27517382 ------CCACAUGGUUUUGGGCUUGGACAAAAGUUAGCCGAAGCGCAAACCACUUGCGUCGGGUAUCACAACAUCCAGACUCUGUCAGUUACUAAC---GCUCUUGCGAAAAUCU ------......(((((((((..((((.......((.(((..((((((.....))))))))).))........))))..))).............(---((....))))))))). ( -25.56, z-score = -0.32, R) >consensus ______CCACAUGGUUUUGGGCUUCGACAAAAGUUAGCCGAAGCGCAAACCACUUGCGUCGGGUGUCACAACACCCAGACCCUGUCAGCUACUAAC___GCCCUUGCGAAAAUCU ............((((.((.((((.(.(........).).)))).))))))..((((...(((((......))))).(((...)))...................))))...... ( -9.61 = -9.73 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:14 2011