
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,493,189 – 2,493,302 |
| Length | 113 |
| Max. P | 0.997514 |

| Location | 2,493,189 – 2,493,281 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Shannon entropy | 0.12870 |
| G+C content | 0.35735 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -17.93 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.05 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
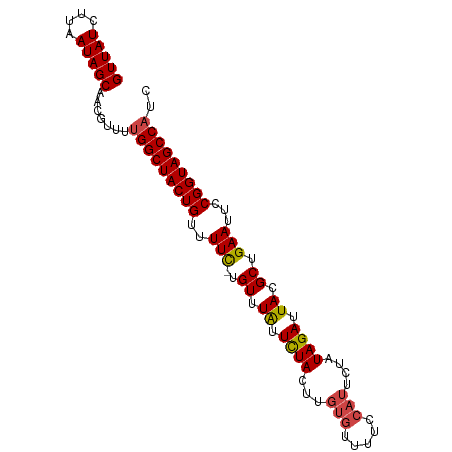
>dm3.chr3R 2493189 92 + 27905053 GUUAUCUUAAUAGCAACGUUUUGGCUACUGUUUUC-UGUUUGUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUC (((((....))))).......(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...))))))))).. ( -22.00, z-score = -3.53, R) >droSim1.chr3R 2525193 92 + 27517382 GUUAUCUUAAUAGCAACGUUUUGGCUACUGUUUUC-UGUUUAUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUC (((((....))))).......(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...))))))))).. ( -22.80, z-score = -4.13, R) >droSec1.super_6 2587424 92 + 4358794 GUUAUCUUAAUAGCAACGUUUUGGCUACUGUUUUC-UGUUUAUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUC (((((....))))).......(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...))))))))).. ( -22.80, z-score = -4.13, R) >droYak2.chr3R 20909162 92 - 28832112 GUUAUCUUAAUAGCAACGUUUUGGCUACUGUUUUC-UGUUUAUUCUACUUGUGUUUUCCAUUCCAUAGAUUACGCUGAAUUCCGGUAGCCAUC (((((....))))).......(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...))))))))).. ( -22.80, z-score = -4.10, R) >droEre2.scaffold_4770 2736253 91 + 17746568 GUUAUCUUAAUAGCAACGUUUUGGCUACUGGUUUC-UGUUUAUUCUACUUGUGUUU-CCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUC (((((....))))).......((((((((((.(((-.((.((.((((...(((...-.)))....)))).)).)).)))..)))))))))).. ( -26.40, z-score = -4.91, R) >droAna3.scaffold_13340 18155655 86 - 23697760 GUUAUCUUAAUAGCAACGUUUUGGCUACUGUUUUUGUGUUUAUUUUAUUU-------CUAUUUUUUAGAUUACGCUGAAUUCCGGUAGCCUUC (((((....)))))........((((((((.(((.((((..........(-------(((.....))))..)))).)))...))))))))... ( -18.90, z-score = -3.48, R) >consensus GUUAUCUUAAUAGCAACGUUUUGGCUACUGUUUUC_UGUUUAUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUC (((((....))))).......(((((((((.......((.((.((((...(((.....)))....)))).)).)).......))))))))).. (-17.93 = -18.32 + 0.39)


| Location | 2,493,203 – 2,493,302 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.87 |
| Shannon entropy | 0.15487 |
| G+C content | 0.36618 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
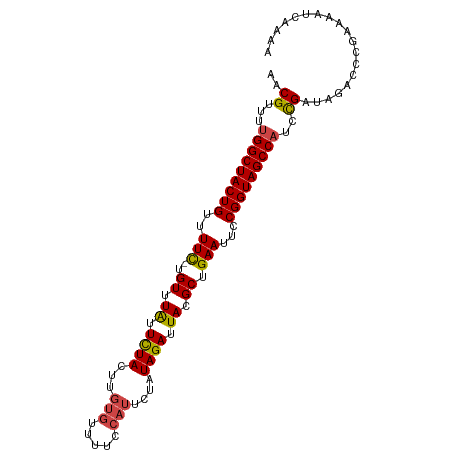
>dm3.chr3R 2493203 99 + 27905053 AACGUUUUGGCUACUGUUUUC-UGUUUGUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUCCGAUAGACCCGAAAAUCAAAG ..((...(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...)))))))))..))................... ( -21.60, z-score = -2.14, R) >droSim1.chr3R 2525207 99 + 27517382 AACGUUUUGGCUACUGUUUUC-UGUUUAUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUCCGAUAGACCCGAAAAUCAAAA ..((...(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...)))))))))..))................... ( -22.40, z-score = -2.82, R) >droSec1.super_6 2587438 99 + 4358794 AACGUUUUGGCUACUGUUUUC-UGUUUAUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUCCGAUAGACCCGAAAAUCAAAA ..((...(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...)))))))))..))................... ( -22.40, z-score = -2.82, R) >droYak2.chr3R 20909176 99 - 28832112 AACGUUUUGGCUACUGUUUUC-UGUUUAUUCUACUUGUGUUUUCCAUUCCAUAGAUUACGCUGAAUUCCGGUAGCCAUCUGAUAGACCCGAAAAUCAAAA ...(((.(((((((((..(((-.((.((.((((...(((.....)))....)))).)).)).)))...)))))))))...)))................. ( -21.20, z-score = -2.36, R) >droEre2.scaffold_4770 2736267 98 + 17746568 AACGUUUUGGCUACUGGUUUC-UGUUUAUUCUACUUGUGUUU-CCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUCUGAUAGACCCGAAAAUCAAAA ...(((.((((((((((.(((-.((.((.((((...(((...-.)))....)))).)).)).)))..))))))))))...)))................. ( -24.80, z-score = -3.07, R) >droAna3.scaffold_13340 18155669 92 - 23697760 AACGUUUUGGCUACUGUUUUUGUGUUUAUUUUAUUU-------CUAUUUUUUAGAUUACGCUGAAUUCCGGUAGCCUUCCGAUAAACUC-AAAAUCAAAA ..((....((((((((.(((.((((..........(-------(((.....))))..)))).)))...))))))))...))........-.......... ( -17.90, z-score = -2.45, R) >consensus AACGUUUUGGCUACUGUUUUC_UGUUUAUUCUACUUGUGUUUUCCAUUCUAUAGAUUACGCUGAAUUCCGGUAGCCAUCCGAUAGACCCGAAAAUCAAAA ..((...(((((((((.......((.((.((((...(((.....)))....)))).)).)).......)))))))))..))................... (-17.29 = -17.46 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:11 2011