| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,469,912 – 2,470,016 |
| Length | 104 |
| Max. P | 0.902386 |

| Location | 2,469,912 – 2,470,016 |
|---|---|
| Length | 104 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 62.47 |
| Shannon entropy | 0.70836 |
| G+C content | 0.51587 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.48 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
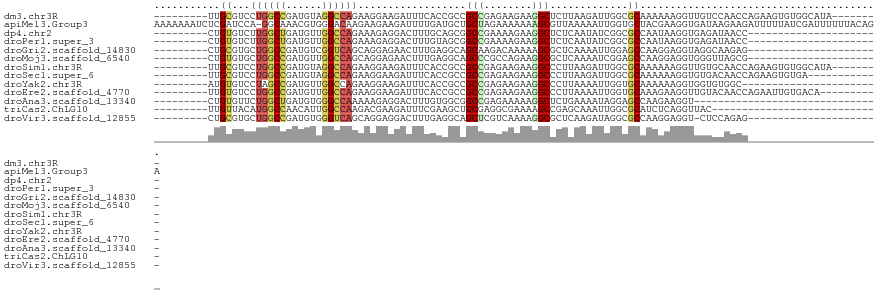
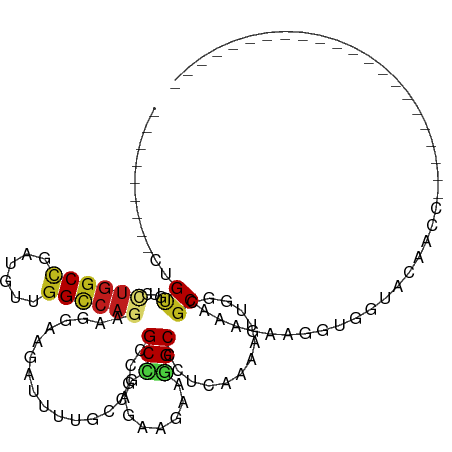
>dm3.chr3R 2469912 104 - 27905053 ---------UUGCGUCCUGGCCGAUGUAGGCCAGAAGGAAGAUUUCACCGCCGCCGAGAAGAAGGCUCUUAAGAUUGGCGCAAAAAAGGUUGUCCAACCAGAAGUGUGGCAUA-------- ---------.(((...((((((......)))))).....(.(((((...((.(((((.((((....))))....)))))))......((((....)))).))))).).)))..-------- ( -33.90, z-score = -1.58, R) >apiMel3.Group3 9821646 120 + 12341916 AAAAAAAUCUCGAUCCA-GGCAAACGUGGGACAAGAAGAAGAUUUUGAUGCUGCUAGAAAAAAAGCGUUAAAAAUUGGUGCUACGAAGGUGAUAAGAAGAUUUUUAUCGAUUUUUUACAGA .((((((((........-.((...(((((.((.......((.((((((((((...........)))))))))).)).)).)))))...))(((((((....)))))))))))))))..... ( -25.70, z-score = -1.36, R) >dp4.chr2 10888077 90 + 30794189 ---------CUGUGUCUUGGCUGAUGUUGGCCAGAAAGAGGACUUUGCAGCGGCCGAAAAGAAGGCUCUCAAUAUCGGCGCCAAUAAGGUGAGAUAACC---------------------- ---------...((((((.((((((((((((...((((....))))...))((((........))))..))))))))))(((.....)))))))))...---------------------- ( -28.10, z-score = -1.23, R) >droPer1.super_3 4723622 90 + 7375914 ---------CUGUGUCUUGGCUGAUGUUGGCCAGAAAGAGGACUUUGUAGCGGCCGAAAAGAAGGCUCUCAAUAUCGGCGCCAAUAAGGUGAGAUAACC---------------------- ---------...((((((.((((((((((((.(.((((....)))).).))((((........))))..))))))))))(((.....)))))))))...---------------------- ( -30.10, z-score = -2.26, R) >droGri2.scaffold_14830 4761265 90 + 6267026 ---------CUGCGUGCUGGCCGAUGUCGGUCAGCAGGAGAACUUUGAGGCAGCAAGACAAAAAGCGCUCAAAAUUGGAGCCAAGGAGGUAGGCAAGAG---------------------- ---------((((.(((((((((....)))))))))......(((((.....((..........))((((.......)))))))))..)))).......---------------------- ( -29.30, z-score = -1.40, R) >droMoj3.scaffold_6540 10025207 90 + 34148556 ---------CUGUGUGCUGGCCGAUGUUGGCCAGCAGGAGAACUUUGAGGCAGCCCGCCAGAAGGCGCUCAAAAUCGGAGCCAAGGAGGUGGGUUAGCG---------------------- ---------((.(.(((((((((....))))))))).))).........((((((((((.....(.((((.......))))).....)))))))).)).---------------------- ( -38.70, z-score = -2.11, R) >droSim1.chr3R 2502147 104 - 27517382 ---------UUGCGUCCUGGCCGAUGUAGGCCAGAAGGAAGAUUUCACCGCCGCCGAGAAGAAGGCCCUUAAGAUUGGCGCAAAAAAGGUUGUGCAACCAGAAGUGUGGCAUA-------- ---------(((((((((((((......))))))((((..(....)......(((........)))))))......))))))).....((..(((........)))..))...-------- ( -33.00, z-score = -1.31, R) >droSec1.super_6 2563044 100 - 4358794 ---------UUGCGUCCUGGCCGAUGUAGGCCAGAAGGAAGAUUUCACCGCCGCCGAGAAGAAGGCCCUUAAGAUUGGCGCAAAAAAGGUGUGACAACCAGAAGUGUGA------------ ---------(((((((((((((......))))))((((..(....)......(((........)))))))......)))))))....(((......)))..........------------ ( -31.60, z-score = -1.94, R) >droYak2.chr3R 20885649 89 + 28832112 ---------AUGUGUCCUAGCCGAUGUUGGCCAGAAGGAAGAUUUCACCGCCGCCGAGAAGAAGGCCCUUAAAAUUGGUGCAAAAAAGGUGGUGUGGC----------------------- ---------.....((((.((((....))))....))))......((((((((((........)))((........)).........)))))))....----------------------- ( -24.90, z-score = -0.63, R) >droEre2.scaffold_4770 2712925 102 - 17746568 ---------UUGUGUCCUGGCCGAUGUUGGCCAGAAGGAAGAUUUCACCGCCGCCGAGAAGAAGGCCCUUAAAAUUGGUGCAAAGAAGGUUGUACAACCAGAAUUGUGACA---------- ---------...(((((((((((....))))))).........(((...((.(((((.(((......)))....)))))))......((((....)))).)))....))))---------- ( -27.20, z-score = -0.80, R) >droAna3.scaffold_13340 13692638 81 - 23697760 ---------CUGUGUUCUGGCUGAUGUGGGCCAAAAAGAGGACUUUGUGGCGGCCGAGAAAAAGGCUCUGAAAAUAGGAGCCAAGAAGGU------------------------------- ---------.....((((((((.......((((.((((....)))).)))))))).))))...((((((.......))))))........------------------------------- ( -24.51, z-score = -1.74, R) >triCas2.ChLG10 3151925 84 + 8806720 ---------UUGUUACAUGGCCAACAUUGGCCAAGACGAAGAUUUCGAAGCUGCGAGGCGAAAAGCCGAGCAAAUUGGCGCAUCUCAGGUUAC---------------------------- ---------(((((...((((((....)))))).))))).........((((..(((..(....(((((.....))))).)..))).))))..---------------------------- ( -24.50, z-score = -1.29, R) >droVir3.scaffold_12855 1179981 89 + 10161210 ---------CUGCGUGCUGGCCGAUGUGGGUCAGCAGGAGGACUUUGAGGCAGCUCGUCAAAAGGCGCUCAAGAUAGGCGCCAAGGAGGU-CUCCAGAG---------------------- ---------.....((((((((......))))))))((((..(((((((....))).......((((((.......))))))..))))..-))))....---------------------- ( -37.50, z-score = -1.96, R) >consensus _________CUGUGUCCUGGCCGAUGUUGGCCAGAAGGAAGAUUUUGCCGCCGCCGAGAAGAAGGCCCUCAAAAUUGGCGCAAAGAAGGUGGUACAACC______________________ ...........((...((((((......))))))..................(((........))).............))........................................ (-11.46 = -11.48 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:08 2011