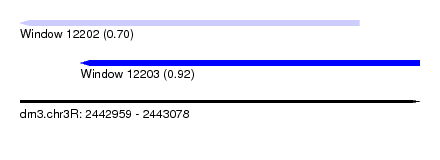
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,442,959 – 2,443,078 |
| Length | 119 |
| Max. P | 0.922799 |

| Location | 2,442,959 – 2,443,060 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Shannon entropy | 0.34428 |
| G+C content | 0.44515 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -16.53 |
| Energy contribution | -18.28 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
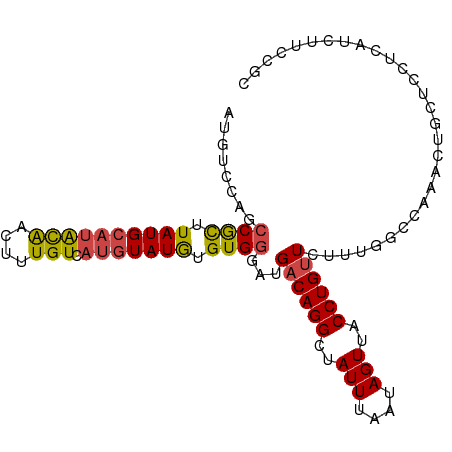
>dm3.chr3R 2442959 101 - 27905053 AUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAACUGCUCCUCAUCUUCCGC .((((...((((.((((((((((....))).))))))).))))...))))(((((...(((((....)))))...)))))..................... ( -26.50, z-score = -1.64, R) >droSim1.chr3R 2472758 101 - 27517382 AUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAACUGCUCCUCAUCUUCCGC .((((...((((.((((((((((....))).))))))).))))...))))(((((...(((((....)))))...)))))..................... ( -26.50, z-score = -1.64, R) >droSec1.super_6 2536473 90 - 4358794 AUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUU-----------CUGCUUCUCAUCUUCCGC ........((((.((((((((((....))).))))))).))))(((((.((((.....(((((....)))))-----------..)))).)))))...... ( -23.60, z-score = -1.81, R) >droYak2.chr3R 18359979 100 - 28832112 -UGUCCAGCCGCUUAUGCAUGCAACUUUGUCAUGUAUGUGUGGAAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCACACGGCUCCUCAUCUUCCGC -((...((((....(((((((((....)).)))))))((((((...((((((..(((....)))..))))))......))))))))))...))........ ( -30.40, z-score = -2.45, R) >droEre2.scaffold_4770 2686484 101 - 17746568 AUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAACCGCUCCUCGUCUUCCGC .((((...((((.((((((((((....))).))))))).))))...))))(((((...(((((....)))))...)))))..................... ( -26.50, z-score = -1.57, R) >droPer1.super_3 4699309 90 + 7375914 AUGCCCUUCCAUGUAUGCGUGUGUGUGUGU---AUAUAUGUGAGUUGACAGGCUAUUUAAUAGUUACCUGCUGUC--------CAGGCAUUUGUCUUUUGU (((((((..((((((((((........)))---)))))))..))..((((((((((...)))))......)))))--------..)))))........... ( -23.00, z-score = -1.46, R) >consensus AUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAACUGCUCCUCAUCUUCCGC .((((...((((.((((((((((....))).))))))).))))...))))(((((...(((((....)))))...)))))..................... (-16.53 = -18.28 + 1.75)

| Location | 2,442,977 – 2,443,078 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.44777 |
| G+C content | 0.43243 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.38 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
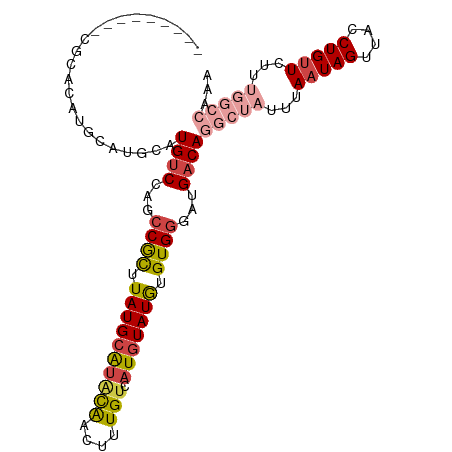
>dm3.chr3R 2442977 101 - 27905053 ----ACACGCGCACAUGAAUGAAUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAA ----...................((((...((((.((((((((((....))).))))))).))))...))))(((((...(((((....)))))...)))))... ( -26.50, z-score = -0.59, R) >droSim1.chr3R 2472776 97 - 27517382 --------CCGCACAUGCAUGCAUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAA --------(((((((((((((((.........((....)).........)).))))))))))))).......(((((...(((((....)))))...)))))... ( -32.97, z-score = -2.45, R) >droSec1.super_6 2536489 88 - 4358794 --------CCGCACAUGCAUGCAUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCU--------- --------(((((((((((((((.........((....)).........)).)))))))))))))...((((((..(((....)))..))))))..--------- ( -30.17, z-score = -2.58, R) >droYak2.chr3R 18359997 82 - 28832112 -----------------------UGUCCAGCCGCUUAUGCAUGCAACUUUGUCAUGUAUGUGUGGAAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCACA -----------------------((((...((((.((((((((((....)).)))))))).))))...))))(((((...(((((....)))))...)))))... ( -29.30, z-score = -3.44, R) >droEre2.scaffold_4770 2686502 105 - 17746568 ACGGCGACACGCAAAUGCAUGCAUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAA .((((((((.(((......))).))))..))))((((((((((((.........))))))))))))......(((((...(((((....)))))...)))))... ( -34.20, z-score = -1.89, R) >droPer1.super_3 4699327 76 + 7375914 ------------------AUACAUGCCCUUCCAUGUAUGCGUGUGUGUGUG---UAUAUAUGUGAGUUGACAGGCUAUUUAAUAGUUACCUGCUGUC-------- ------------------(((((((......)))))))(((((((((....---))))))))).....((((((((((...)))))......)))))-------- ( -16.40, z-score = 0.04, R) >consensus _________CGCACAUGCAUGCAUGUCCAGCCGCUUAUGCAUACAACUUUGUCAUGUAUGUGUGGGAUGACAGGCUAUUUAAUAGUUACCUGUUCUUUGGCCAAA .......................((((...((((.((((((((((....))).))))))).))))...))))(((((...(((((....)))))...)))))... (-16.66 = -18.38 + 1.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:07 2011