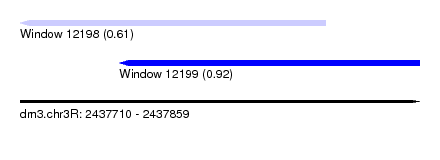
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,437,710 – 2,437,859 |
| Length | 149 |
| Max. P | 0.921091 |

| Location | 2,437,710 – 2,437,824 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Shannon entropy | 0.47244 |
| G+C content | 0.53832 |
| Mean single sequence MFE | -39.64 |
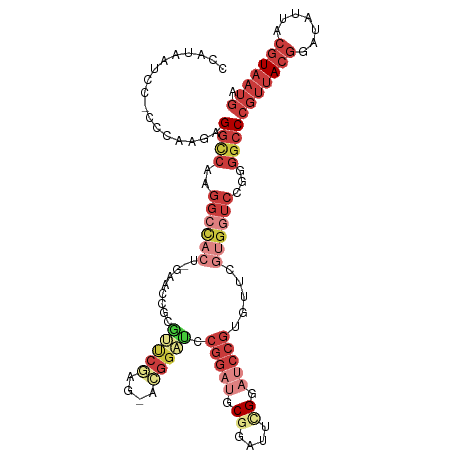
| Consensus MFE | -19.35 |
| Energy contribution | -22.13 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
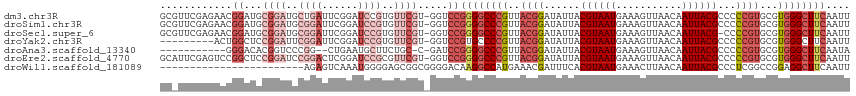
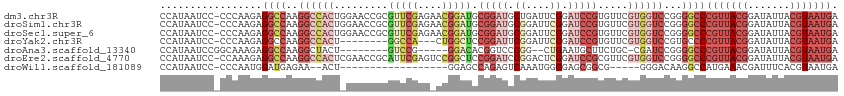
>dm3.chr3R 2437710 114 - 27905053 GCGUUCGAGAACGGAUGCGGAUGCUGAUUCGGAUCCGUGUUCGU-GGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUU ......((..((((((((((((.(......).))))))))))))-..)).(((((((((((((.......))))))..............((((......))))))))))).... ( -41.90, z-score = -1.31, R) >droSim1.chr3R 2467834 114 - 27517382 GCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGU-GGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUU ..(((((...((((((((((((((((((....))))))))))))-.))))(((((.(((((((.......)))))))...((........))))))).))...)))))....... ( -43.90, z-score = -1.45, R) >droSec1.super_6 2531614 113 - 4358794 GCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGU-GGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACG-CCCCGUGCGUGGGCUUCAAUU ..(((((...((((((((((((.((....)).))))))))))))-.(..((((((.(((((((.......)))))))...((........)))-)))))..).)))))....... ( -45.60, z-score = -2.15, R) >droYak2.chr3R 18354665 105 - 28832112 ---------ACUGGCUCCGGAUUCGGAUUCGGAUCCGUGUUCGU-GGUCCGUGCCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUU ---------((.(((..(((((.(((((....))))).))))).-.))).))(((((((.((((......((((((...........))))))...))))))).))))....... ( -36.70, z-score = -1.60, R) >droAna3.scaffold_13340 13664340 100 - 23697760 -----------GGGACACGGUCCCGG--CUGAAUGCUUCUGC-C-GAUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUA -----------(((((...)))))..--.((((.(((..(((-(-(....(((((.(((((((.......)))))))...((........))))))))).)))..)))))))... ( -35.80, z-score = -1.16, R) >droEre2.scaffold_4770 2681361 114 - 17746568 GCAUUCGAGUCCGGCUCCGGAUCCGGACUCGGAUCCGCGUUCGU-GGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUU ..(((.((((((((((((((((((((((.((....)).))))).-)))))))))))(((((((.......))))))).............((((......)))))))))).))). ( -51.20, z-score = -3.34, R) >droWil1.scaffold_181089 670541 91 - 12369635 ------------------------AGAGUCAAAUGGGGAGCGGCGGGGACAAGGCCAUGAAACGAUUUCACGUAAUGAAACUUAACAAUUACGCCCUCGGCCGGAGGCUUCAAUU ------------------------............(((((..((....)..((((.((((.....))))((((((...........)))))).....)))).)..))))).... ( -22.40, z-score = 0.05, R) >consensus GC_UUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGU_GGUCCGGGGCCCGUUACGGAUAUUACGUAAUGAAAGUUAACAAUUACGCCCCCGUGCGUGGGCUUCAAUU ..................((((.(((((.((....)).)))))...))))((((((((..((((......((((((...........))))))...))))...)))))))).... (-19.35 = -22.13 + 2.78)
| Location | 2,437,747 – 2,437,859 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.39 |
| Shannon entropy | 0.52534 |
| G+C content | 0.57131 |
| Mean single sequence MFE | -41.93 |
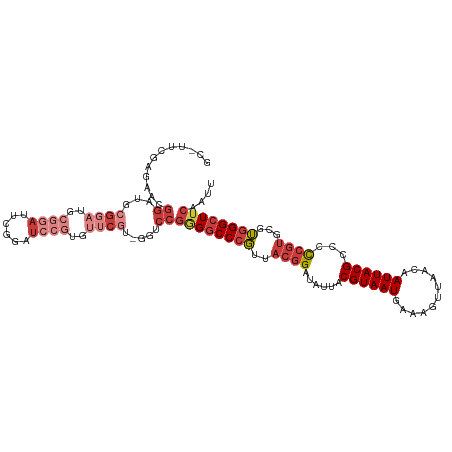
| Consensus MFE | -18.77 |
| Energy contribution | -22.54 |
| Covariance contribution | 3.77 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.921091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2437747 112 - 27905053 CCAUAAUCC-CCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCUGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGA ........(-(((.((.(((....))).))(((.(((((.......((((((.(((((....))))).))))))...))))))))))))..(((((((.......))))))). ( -47.50, z-score = -2.64, R) >droSim1.chr3R 2467871 112 - 27517382 CCAUAAUCC-CCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGA ........(-(((.((.(((....))).))(((.(((((((....)))((((((((((.((....)).)))))))))))))))))))))..(((((((.......))))))). ( -49.40, z-score = -2.70, R) >droSec1.super_6 2531650 112 - 4358794 CCAUAAUCC-CCCAAGAGGCCAAGGCCACUGGAACCGCGUUCGAGAACGGAUGCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGA ........(-(((.((.(((....))).))(((.(((((((....)))((((((((((.((....)).)))))))))))))))))))))..(((((((.......))))))). ( -49.40, z-score = -2.70, R) >droYak2.chr3R 18354702 101 - 28832112 CCAUAAUCC-CCCAAGAGGCCAAGGCCACU--------GGCCA---CUGGCUCCGGAUUCGGAUUCGGAUCCGUGUUCGUGGUCCGUGCCCCGUUACGGAUAUUACGUAAUGA .........-.......(((....)))((.--------(((((---(..((..((((((((....)))))))).))..)))))).))....(((((((.......))))))). ( -37.80, z-score = -1.62, R) >droAna3.scaffold_13340 13664377 97 - 23697760 CCAUAAUCCGGCAAAGAGGCCAAGGCUACU--------GUCCG-----GGACACGGUCCCGG--CUGAAUGCUUCUGC-CGAUCCGGGGCCCGUUACGGAUAUUACGUAAUGA ......(((((...((.(((....))).))--------..)))-----))....((((((((--......((....))-....))))))))(((((((.......))))))). ( -37.50, z-score = -1.49, R) >droEre2.scaffold_4770 2681398 112 - 17746568 CCAUAAUCC-CCAAAGAGGCCAAGGCCACUCGAACCGCAUUCGAGUCCGGCUCCGGAUCCGGACUCGGAUCCGCGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGA .........-.......(((....)))(((((((.....)))))))..((((((((((((((((.((....)).))))).)))))))))))(((((((.......))))))). ( -52.50, z-score = -3.77, R) >droWil1.scaffold_181089 670578 87 - 12369635 CCAUAAUCC-CCCAAUGGAUGAGAA--ACU------------------GGAGCCAGAGUCAAAUGGGGAGCGGCG-----GGGACAAGGCCAUGAAACGAUUUCACGUAAUGA .(((....(-((((...(((.....--.((------------------(....))).)))...)))))...((((-----....)...))))))..(((......)))..... ( -19.40, z-score = 0.93, R) >consensus CCAUAAUCC_CCCAAGAGGCCAAGGCCACU_GAACCGCGUUCGAG_ACGGAUCCGGAUGCGGAUUCGGAUCCGUGUUCGUGGUCCGGGGCCCGUUACGGAUAUUACGUAAUGA .................((((..((((((.........(((((....))))).(((((.((....)).))))).....))))))...))))(((((((.......))))))). (-18.77 = -22.54 + 3.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:03 2011