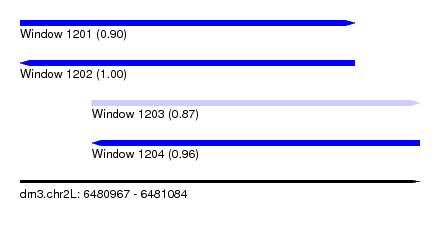
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,480,967 – 6,481,084 |
| Length | 117 |
| Max. P | 0.998390 |

| Location | 6,480,967 – 6,481,065 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.37 |
| Shannon entropy | 0.11589 |
| G+C content | 0.45918 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
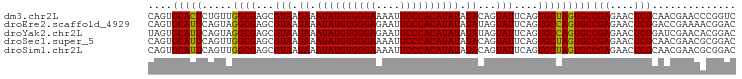
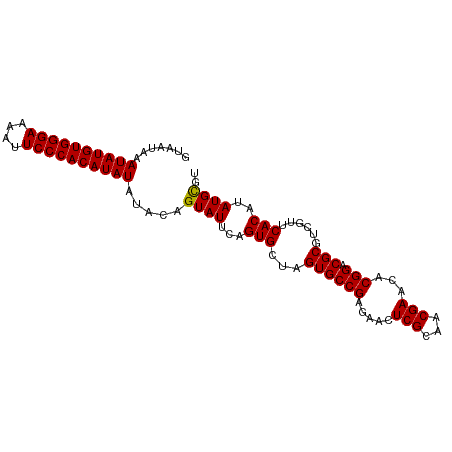
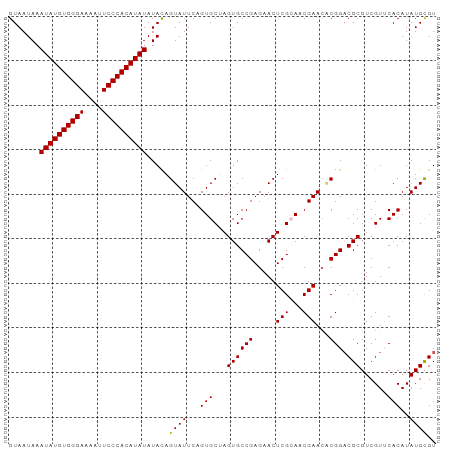
>dm3.chr2L 6480967 98 + 23011544 CAGUGCACUCUGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACCCGGUC ..........(((((.((((........((((((((((....)))))))))).((((((((...))))).))).......)))))))))......... ( -26.50, z-score = -1.37, R) >droEre2.scaffold_4929 15389837 98 + 26641161 CAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAGAAUUCCCACAUAUAUAUAGUAUUCAGUGCCAGUGCCGAGAACUCGGACCGAAAACGGAC ..............((((((..(.(((.(((((((((......))))))))).))).)..)))...)))....((((....)))).(((....))).. ( -28.30, z-score = -2.70, R) >droYak2.chr2L 15888557 98 - 22324452 UAGUGCAUUCAGUAGGCGAGCGUAAUAAAUAUGUGGGAGAAUUCCCACAUAUAUAUAGUAUUCAGUGCCAGUGCCGAGAACUCGGAUCGAACACGGAC ..(((..(((....((((((..(.(((.(((((((((......))))))))).))).)..)))...)))....((((....))))...)))))).... ( -29.10, z-score = -2.76, R) >droSec1.super_5 4530271 98 + 5866729 CAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACGCGGAC ...(((.(((.((((.((((........((((((((((....)))))))))).((((((((...))))).))).......))))))))))).)))... ( -30.80, z-score = -2.91, R) >droSim1.chr2L 6272120 98 + 22036055 CAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACGCGGAC ...(((.(((.((((.((((........((((((((((....)))))))))).((((((((...))))).))).......))))))))))).)))... ( -30.80, z-score = -2.91, R) >consensus CAGUGCAUUCAGUUGGCGAGCGUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACACGGAC ....(((((.....((((...(((.((.((((((((((....)))))))))).))...)))....)))))))))(((....))).............. (-22.90 = -22.50 + -0.40)
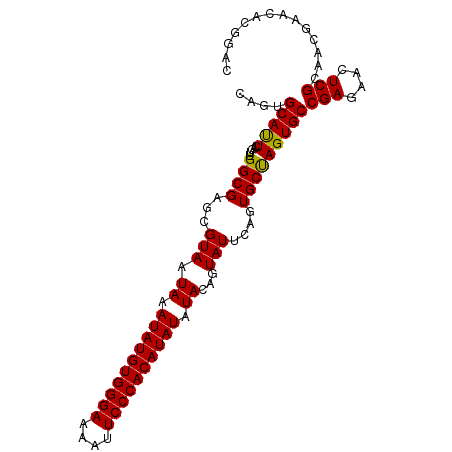
| Location | 6,480,967 – 6,481,065 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Shannon entropy | 0.11589 |
| G+C content | 0.45918 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -28.82 |
| Energy contribution | -29.86 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.08 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6480967 98 - 23011544 GACCGGGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACAGAGUGCACUG ....(.(((((((((((((((...((....))...)).....(((.((((((((((....))))))))))....)))))))).)))).)))).).... ( -30.30, z-score = -2.53, R) >droEre2.scaffold_4929 15389837 98 - 26641161 GUCCGUUUUCGGUCCGAGUUCUCGGCACUGGCACUGAAUACUAUAUAUAUGUGGGAAUUCUCCCACAUAUUUAUUACGCUCGCCUACUGAAUGCACUG ....((.((((((((((....))))....(((...((.....(((.((((((((((....)))))))))).))).....))))).)))))).)).... ( -29.80, z-score = -3.61, R) >droYak2.chr2L 15888557 98 + 22324452 GUCCGUGUUCGAUCCGAGUUCUCGGCACUGGCACUGAAUACUAUAUAUAUGUGGGAAUUCUCCCACAUAUUUAUUACGCUCGCCUACUGAAUGCACUA ....(((((((..((((....))))....(((...((.....(((.((((((((((....)))))))))).))).....)))))...))))))).... ( -28.90, z-score = -3.22, R) >droSec1.super_5 4530271 98 - 5866729 GUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUG ....(((((((((((((((((...((....))...)).....(((.((((((((((....))))))))))....)))))))).)))).)))))).... ( -37.10, z-score = -5.51, R) >droSim1.chr2L 6272120 98 - 22036055 GUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUG ....(((((((((((((((((...((....))...)).....(((.((((((((((....))))))))))....)))))))).)))).)))))).... ( -37.10, z-score = -5.51, R) >consensus GUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUACGCUCGCCAACUGAAUGCACUG ....(((((((((((((((((...((....))...)).....(((.((((((((((....)))))))))).)))...))))).)))).)))))).... (-28.82 = -29.86 + 1.04)
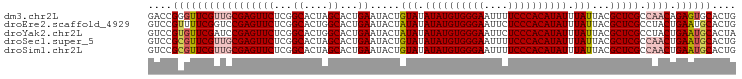
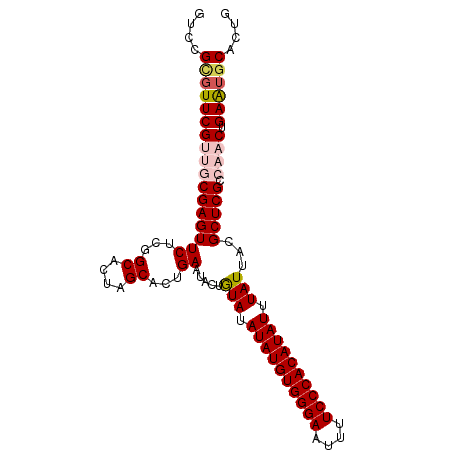
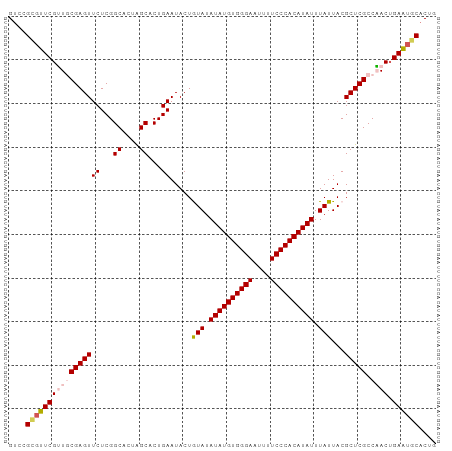
| Location | 6,480,988 – 6,481,084 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Shannon entropy | 0.10326 |
| G+C content | 0.45000 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6480988 96 + 23011544 GUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACCCGGUCGCGUCGUUCACAUAUGUGU .......((((((((((....))))))))))(((((.......(.((....)))..((((.(((..((....))...)))..)))).....))))) ( -26.30, z-score = -2.05, R) >droEre2.scaffold_4929 15389858 96 + 26641161 GUAAUAAAUAUGUGGGAGAAUUCCCACAUAUAUAUAGUAUUCAGUGCCAGUGCCGAGAACUCGGACCGAAAACGGACGCGUCGUGCACAUAUGCGU ...(((.(((((((((......))))))))).))).((((...(((((.(((((((....)))).(((....))).)))...).))))..)))).. ( -32.20, z-score = -3.29, R) >droYak2.chr2L 15888578 96 - 22324452 GUAAUAAAUAUGUGGGAGAAUUCCCACAUAUAUAUAGUAUUCAGUGCCAGUGCCGAGAACUCGGAUCGAACACGGACGCGUCGUGCACAUAUGCGU ((((((.(((((((((......))))))))).))).((((...(((((.(((((((....))))......))))).)))...)))).....))).. ( -29.80, z-score = -2.16, R) >droSec1.super_5 4530292 96 + 5866729 GUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACGCGGACGCGUCGUUCACAUAUGCGU .......((((((((((....)))))))))).....((((...(((.....((.((....))))(((((.(((....)))))))))))..)))).. ( -28.60, z-score = -2.21, R) >droSim1.chr2L 6272141 96 + 22036055 GUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACGCGGACGCGUCGUUCACAUAUGCGU .......((((((((((....)))))))))).....((((...(((.....((.((....))))(((((.(((....)))))))))))..)))).. ( -28.60, z-score = -2.21, R) >consensus GUAAUAAAUAUGUGGGAAAAUUCCCACAUAUAUACAGUAUUCAGUGCUAGUGCCGAGAACUCGCAACGAACACGGACGCGUCGUUCACAUAUGCGU .......((((((((((....)))))))))).....((((...(((...((((((.....(((...)))...))).)))......)))..)))).. (-23.46 = -23.30 + -0.16)
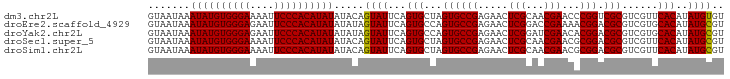
| Location | 6,480,988 – 6,481,084 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Shannon entropy | 0.10326 |
| G+C content | 0.45000 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -24.82 |
| Energy contribution | -24.66 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
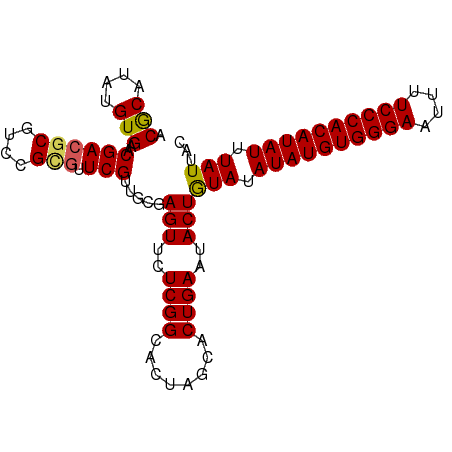
>dm3.chr2L 6480988 96 - 23011544 ACACAUAUGUGAACGACGCGACCGGGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUAC .(((....))).....((((((.......))))))(((..((((........))))..)))(((.((((((((((....)))))))))).)))... ( -28.50, z-score = -2.54, R) >droEre2.scaffold_4929 15389858 96 - 26641161 ACGCAUAUGUGCACGACGCGUCCGUUUUCGGUCCGAGUUCUCGGCACUGGCACUGAAUACUAUAUAUAUGUGGGAAUUCUCCCACAUAUUUAUUAC ........((((..((((....))))..((((((((....)))).))))))))........(((.((((((((((....)))))))))).)))... ( -30.80, z-score = -3.07, R) >droYak2.chr2L 15888578 96 + 22324452 ACGCAUAUGUGCACGACGCGUCCGUGUUCGAUCCGAGUUCUCGGCACUGGCACUGAAUACUAUAUAUAUGUGGGAAUUCUCCCACAUAUUUAUUAC ..((....((((.((((((....))).)))...(((....)))))))..))..........(((.((((((((((....)))))))))).)))... ( -30.60, z-score = -2.77, R) >droSec1.super_5 4530292 96 - 5866729 ACGCAUAUGUGAACGACGCGUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUAC .((((....((((((.((....)))))))).))))(((..((((........))))..)))(((.((((((((((....)))))))))).)))... ( -31.00, z-score = -2.80, R) >droSim1.chr2L 6272141 96 - 22036055 ACGCAUAUGUGAACGACGCGUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUAC .((((....((((((.((....)))))))).))))(((..((((........))))..)))(((.((((((((((....)))))))))).)))... ( -31.00, z-score = -2.80, R) >consensus ACGCAUAUGUGAACGACGCGUCCGCGUUCGUUGCGAGUUCUCGGCACUAGCACUGAAUACUGUAUAUAUGUGGGAAUUUUCCCACAUAUUUAUUAC .(((....)))..((((((....))).))).....(((..((((........))))..)))(((.((((((((((....)))))))))).)))... (-24.82 = -24.66 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:27 2011