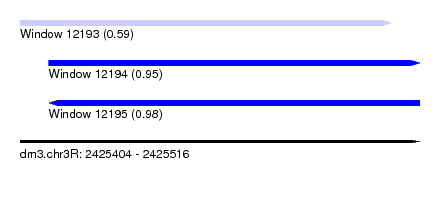
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,425,404 – 2,425,516 |
| Length | 112 |
| Max. P | 0.975450 |

| Location | 2,425,404 – 2,425,508 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.67244 |
| G+C content | 0.39551 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -6.51 |
| Energy contribution | -6.59 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
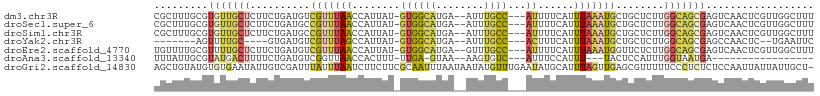
>dm3.chr3R 2425404 104 + 27905053 CGCUUUGCGUGUUGCUCUUCUGAUGUCGUUUAACCAUUAU-GUGGCAUGA--AUUUGCC---AUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUU .(((..(((.(((((((..........((((((.......-((((((...--...))))---))......))))))(((((.....)))))))).)))).))).)))... ( -34.52, z-score = -3.74, R) >droSec1.super_6 2519216 104 + 4358794 CGCUUUGCGUGUUGCUCUUCUGAUGCCGUUUAACCAUUAU-GUGGCAUGA--AUUUGCC---AUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUU .(((..(((.(((((((..........((((((.......-((((((...--...))))---))......))))))(((((.....)))))))).)))).))).)))... ( -34.52, z-score = -3.43, R) >droSim1.chr3R 2454998 104 + 27517382 CGCUUUGCGUGUUGCUCUUCUGAUGCCGUUUAACCAUUAU-GUGGCAUGA--AUUUGCC---AUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUU .(((..(((.(((((((..........((((((.......-((((((...--...))))---))......))))))(((((.....)))))))).)))).))).)))... ( -34.52, z-score = -3.43, R) >droYak2.chr3R 18341672 91 + 28832112 -------AGUUUUGC----GUGAUGUCGUUUAGCCAUUAU-GUGGCAUGA--AUUUGCC---ACUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGCCAACUC--UGAAUUC -------.(((..((----(......)))..)))......-((((((...--...))))---)).((((.......(((((.....)))))(((....)))--))))... ( -25.70, z-score = -1.65, R) >droEre2.scaffold_4770 2668197 104 + 17746568 UGUUUUGCGUUUUGCUCUUCUGAUGUCGUUUAACCAUUAU-GUGGCAUGA--GUUUGCC---AUUUUCAUUUAAAUGGUUCUCUUGGCAGCGAGUCAACUCGUUGGCUUU ......((.....)).......((((((..((.....)).-.))))))((--(...(((---((((......))))))).)))..(.(((((((....))))))).)... ( -25.80, z-score = -1.52, R) >droAna3.scaffold_13340 13653739 83 + 23697760 UUUAUUGCGUAUGACUUUUCUGAUGUCGGUUAACCACUUU-UUGA-GUAA--AAGUGUC---AUUUCCAUUU---UACUCCAUUUGGUAAUGA----------------- .(((((((....(((.........)))((.....((((((-(...-..))--)))))..---....))....---...........)))))))----------------- ( -13.70, z-score = -0.66, R) >droGri2.scaffold_14830 4714654 109 - 6267026 AGCUGUAUGUGUGAAUAUUGUCGAUUUAUUUAAUCUUCUUCGCAAUUUAAUAAUAUGUUUGAAUAUGCAUUUAGUUGAGCGUUUUUCCCUCUCUCCAAUUAUUAUUGCU- ((((((((.((((((....(..((((.....))))..)))))))((((((........)))))))))))..((((((((.(.........).)).)))))).....)))- ( -12.80, z-score = 0.56, R) >consensus CGCUUUGCGUGUUGCUCUUCUGAUGUCGUUUAACCAUUAU_GUGGCAUGA__AUUUGCC___AUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUU .........(((((((.....(....(((((((..........((((........))))...........)))))))...)....))))))).................. ( -6.51 = -6.59 + 0.07)
| Location | 2,425,412 – 2,425,516 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.51959 |
| G+C content | 0.40256 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
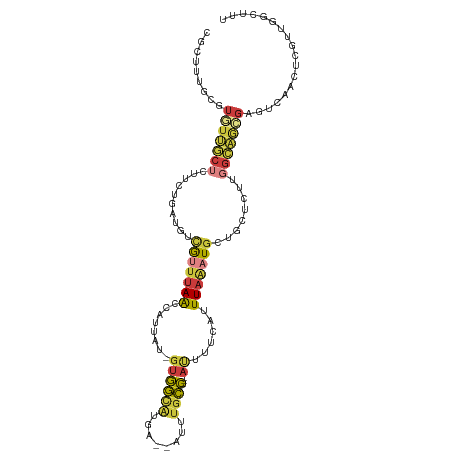
>dm3.chr3R 2425412 104 + 27905053 GUGUUGCUCUUCUGAUGUCGUUUAACCAUU------AUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAC ..(((((((..........((((((.....------..((((((......))))))......))))))(((((.....)))))))).))))(((((((....))))))). ( -30.32, z-score = -2.49, R) >droSec1.super_6 2519224 104 + 4358794 GUGUUGCUCUUCUGAUGCCGUUUAACCAUU------AUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAA ..(((((((..........((((((.....------..((((((......))))))......))))))(((((.....)))))))).))))(((((((....))))))). ( -30.62, z-score = -2.28, R) >droSim1.chr3R 2455006 104 + 27517382 GUGUUGCUCUUCUGAUGCCGUUUAACCAUU------AUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAA ..(((((((..........((((((.....------..((((((......))))))......))))))(((((.....)))))))).))))(((((((....))))))). ( -30.62, z-score = -2.28, R) >droYak2.chr3R 18341680 91 + 28832112 -----------GUGAUGUCGUUUAGCCAUU------AUGUGGCAUGAAUUUGCCACUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGCCAACUC--UGAAUUCCUAGGGAA -----------((((((.(.....).))))------))((((((......))))))...........((((((.....))))))......(((--((......))))).. ( -25.90, z-score = -1.38, R) >droEre2.scaffold_4770 2668205 104 + 17746568 GUUUUGCUCUUCUGAUGUCGUUUAACCAUU------AUGUGGCAUGAGUUUGCCAUUUUCAUUUAAAUGGUUCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUUGCGAU ...((((.......((((((..((.....)------)..))))))(((...(((((((......))))))).)))..(.(((((((....))))))).)......)))). ( -28.30, z-score = -2.33, R) >droGri2.scaffold_14830 4714662 109 - 6267026 GUGUGAAUAUUGUCGAUUUAUUUAAUCUUCUUCGCAAUUUAAUAAUAUGUUUGAAUAUGCAUUUAGUUGAGCGUUUUUCCCUCUCUCCAA-UUAUUAUUGCUCUGUUGAC ...........((((((................(((((((((........)))))).)))........(((((.................-.......))))).)))))) ( -13.36, z-score = 0.01, R) >consensus GUGUUGCUCUUCUGAUGUCGUUUAACCAUU______AUGUGGCAUGAAUUUGCCAUUUUCAUUUAAAUGCUGCUCUUGGCAGCGAGUCAACUCGUUGGCUUUUUAGCGAA ............((((......................((((((......))))))............(((((.....)))))..))))..((((((......)))))). (-14.34 = -14.52 + 0.18)
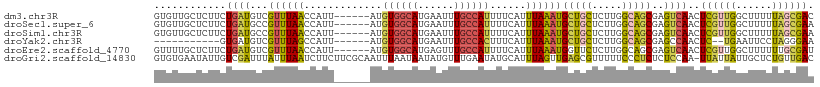
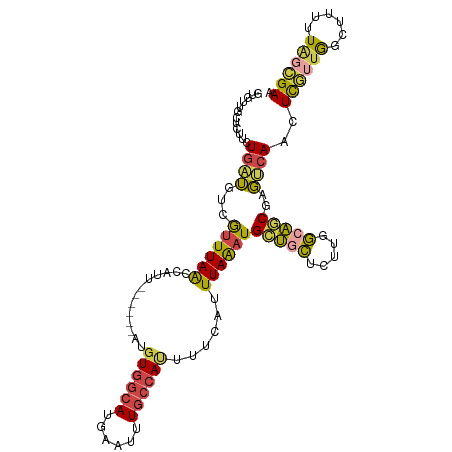
| Location | 2,425,412 – 2,425,516 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.51959 |
| G+C content | 0.40256 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -12.51 |
| Energy contribution | -12.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
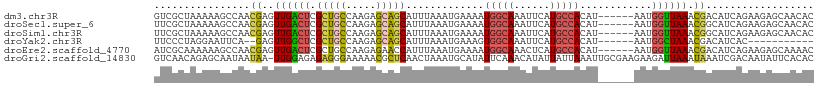
>dm3.chr3R 2425412 104 - 27905053 GUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAU------AAUGGUUAAACGACAUCAGAAGAGCAACAC ((((......(((((..(((....)))(((((.....))))).............(((((......)))))...------..)))))...))))................ ( -25.50, z-score = -2.34, R) >droSec1.super_6 2519224 104 - 4358794 UUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAU------AAUGGUUAAACGGCAUCAGAAGAGCAACAC ...(((.....((((..(((....)))(((((.....))))).............))))...((((((((....------...........)))))..))).)))..... ( -26.46, z-score = -2.24, R) >droSim1.chr3R 2455006 104 - 27517382 UUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAU------AAUGGUUAAACGGCAUCAGAAGAGCAACAC ...(((.....((((..(((....)))(((((.....))))).............))))...((((((((....------...........)))))..))).)))..... ( -26.46, z-score = -2.24, R) >droYak2.chr3R 18341680 91 - 28832112 UUCCCUAGGAAUUCA--GAGUUGGCUCGCUGCCAAGAGCAGCAUUUAAAUGAAAGUGGCAAAUUCAUGCCACAU------AAUGGCUAAACGACAUCAC----------- ((((...))))....--...((((((.(((((.....)))))............((((((......))))))..------...))))))..........----------- ( -24.50, z-score = -1.85, R) >droEre2.scaffold_4770 2668205 104 - 17746568 AUCGCAAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGAACCAUUUAAAUGAAAAUGGCAAACUCAUGCCACAU------AAUGGUUAAACGACAUCAGAAGAGCAAAAC ...((......(.((.((((....)))).)).).....(((((((...(((....(((((......))))))))------)))))))................))..... ( -20.20, z-score = -1.71, R) >droGri2.scaffold_14830 4714662 109 + 6267026 GUCAACAGAGCAAUAAUAA-UUGGAGAGAGGGAAAAACGCUCAACUAAAUGCAUAUUCAAACAUAUUAUUAAAUUGCGAAGAAGAUUAAAUAAAUCGACAAUAUUCACAC (((......((((((((((-(((....(((.(.....).))).......((......))..)).)))))))..))))......((((.....)))))))........... ( -13.80, z-score = -1.25, R) >consensus UUCGCUAAAAAGCCAACGAGUUGACUCGCUGCCAAGAGCAGCAUUUAAAUGAAAAUGGCAAAUUCAUGCCACAU______AAUGGUUAAACGACAUCAGAAGAGCAACAC ...................((((....(((((.....))))).............(((((......)))))...................))))................ (-12.51 = -12.40 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:54:00 2011