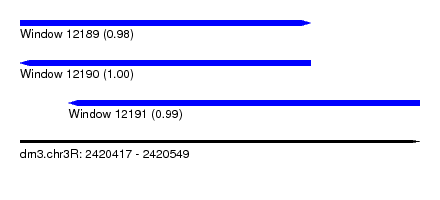
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,420,417 – 2,420,549 |
| Length | 132 |
| Max. P | 0.999940 |

| Location | 2,420,417 – 2,420,513 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 64.40 |
| Shannon entropy | 0.69894 |
| G+C content | 0.42943 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2420417 96 + 27905053 ----------------------AAUGGGAAUCGGGAGGGGUGCUGCAACAUACGUAUUCGUAUACGUA-CUGUACUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC ----------------------..(((...((((.(.((((......)).(((((((....)))))))-)).).))))...)))(((((((((........)))))))))......... ( -23.00, z-score = -1.69, R) >droSim1.chr3R 2448241 96 + 27517382 ----------------------AAUGGGAAUCGGGAGGGGUGCUGCAACAUACGUAUUCGUAUACGUA-CUGUACUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC ----------------------..(((...((((.(.((((......)).(((((((....)))))))-)).).))))...)))(((((((((........)))))))))......... ( -23.00, z-score = -1.69, R) >droSec1.super_6 2514449 96 + 4358794 ----------------------AAUGGGAAUCGGGAGGGGUGCUGCAACAUACGUAUUCGUAUACGUA-CUGUACUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC ----------------------..(((...((((.(.((((......)).(((((((....)))))))-)).).))))...)))(((((((((........)))))))))......... ( -23.00, z-score = -1.69, R) >droYak2.chr3R 18336717 110 + 28832112 --AAUUGGGAAUAGGGUGGAGGGGUGGAGGGGUGGAGAGGUGCUGCAACAUA------CGUAUACGUA-CUGUACUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC --............((((.((..(((..(.(((........))).)..)))(------((....))).-)).)))).(((((..(((((((((........)))))))))..))))).. ( -25.30, z-score = -0.79, R) >droEre2.scaffold_4770 2663107 88 + 17746568 ----------------------AAUGAGAAUCGGG--GGGUGCUGCAACAUA------CGUAUACGUA-CUGUACUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC ----------------------..(((...(((((--.((((((((......------.)))...)))-)).).))))...)))(((((((((........)))))))))......... ( -20.70, z-score = -1.95, R) >dp4.chr2 10835674 77 - 30794189 -----------------------------------ACGGACG-AACGGCAUA----UACG-GUAGGGA-CUGUAGUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC -----------------------------------.((....-..)).(((.----((((-((....)-))))))))(((((..(((((((((........)))))))))..))))).. ( -20.30, z-score = -3.07, R) >droPer1.super_3 4671174 93 - 7375914 -------------------ACGGACGAACGGAGGAACAGCCG-UACAGCAUA----UACG-GUAGGGA-CUGUAGUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC -------------------.((......)).....((((((.-(((.(....----..).-))).)).-))))....(((((..(((((((((........)))))))))..))))).. ( -24.10, z-score = -2.59, R) >droWil1.scaffold_181089 647007 96 + 12369635 ---------------------AAGUGGAAGUGGAAGCGGUAGUUCUAUG--GGAUGUGAAGGAAACUGACUGUAGUGAAAAUCAAUUGAAACGCCGGCAAACGCUUCAAUCAAUUUUAC ---------------------.....((((((...(((((..(((....--.(((....((....)).((....))....)))....)))..))).))...))))))............ ( -19.30, z-score = -0.14, R) >droVir3.scaffold_12855 1104153 79 - 10161210 ---------------------------------------GAAACUUGGGAUGAAAGUGCGAGCAAAUG-CCGUAGUGAAAAUCAAUUGGUGCGCCAACAAACGCUUCAAUCAAUUUUAC ---------------------------------------.(((.(((((((...(.((((.((....)-))))).)....)))..((((.(((........))).)))))))).))).. ( -15.00, z-score = 0.14, R) >droMoj3.scaffold_6540 9973595 118 - 34148556 GACAGAGGCCAGGCCAACUCGAAGCUAGAGGCUACGUAGAGAUUUUAGGUUGUAAGUGCGGGCAAAUG-CAAUAGUGCAAAUCAAUUGGCGCGCCAACAAACGCUUCAAUCAAUUUUAC ((..(((((...(((((((((.(((.....))).)).)).(((((...((((((..((....))..))-)))).....)))))..)))))(......)....)))))..))........ ( -26.30, z-score = 0.70, R) >consensus ______________________AAUGGGAAUCGGGAGGGGUGCUGCAACAUA____UGCGUAUACGUA_CUGUACUGAAAAUCAAUUGAAGCGCCAACAAACGCUUCAAUCAAUUUUAC .............................................................................(((((..(((((((((........)))))))))..))))).. (-11.53 = -11.77 + 0.24)
| Location | 2,420,417 – 2,420,513 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.40 |
| Shannon entropy | 0.69894 |
| G+C content | 0.42943 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -18.37 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2420417 96 - 27905053 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAG-UACGUAUACGAAUACGUAUGUUGCAGCACCCCUCCCGAUUCCCAUU---------------------- (.(((((..(((((((((((....)))))))))))..)))))).(((..(-(((((((....))))))))..)))......................---------------------- ( -30.30, z-score = -4.81, R) >droSim1.chr3R 2448241 96 - 27517382 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAG-UACGUAUACGAAUACGUAUGUUGCAGCACCCCUCCCGAUUCCCAUU---------------------- (.(((((..(((((((((((....)))))))))))..)))))).(((..(-(((((((....))))))))..)))......................---------------------- ( -30.30, z-score = -4.81, R) >droSec1.super_6 2514449 96 - 4358794 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAG-UACGUAUACGAAUACGUAUGUUGCAGCACCCCUCCCGAUUCCCAUU---------------------- (.(((((..(((((((((((....)))))))))))..)))))).(((..(-(((((((....))))))))..)))......................---------------------- ( -30.30, z-score = -4.81, R) >droYak2.chr3R 18336717 110 - 28832112 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAG-UACGUAUACG------UAUGUUGCAGCACCUCUCCACCCCUCCACCCCUCCACCCUAUUCCCAAUU-- (.(((((..(((((((((((....)))))))))))..)))))).(((..(-((((....))------)))..)))..........................................-- ( -27.10, z-score = -4.73, R) >droEre2.scaffold_4770 2663107 88 - 17746568 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAG-UACGUAUACG------UAUGUUGCAGCACCC--CCCGAUUCUCAUU---------------------- (.(((((..(((((((((((....)))))))))))..)))))).(((..(-((((....))------)))..))).......--.............---------------------- ( -27.10, z-score = -4.14, R) >dp4.chr2 10835674 77 + 30794189 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAG-UCCCUAC-CGUA----UAUGCCGUU-CGUCCGU----------------------------------- (((((((..(((((((((((....)))))))))))..))))).))....(-..(..((-.((.----...)).)).-.)..)..----------------------------------- ( -21.80, z-score = -3.52, R) >droPer1.super_3 4671174 93 + 7375914 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAG-UCCCUAC-CGUA----UAUGCUGUA-CGGCUGUUCCUCCGUUCGUCCGU------------------- (((((((..(((((((((((....)))))))))))..))))).)).((((-.((.(((-.((.----...)).)))-.))))))................------------------- ( -29.60, z-score = -4.38, R) >droWil1.scaffold_181089 647007 96 - 12369635 GUAAAAUUGAUUGAAGCGUUUGCCGGCGUUUCAAUUGAUUUUCACUACAGUCAGUUUCCUUCACAUCC--CAUAGAACUACCGCUUCCACUUCCACUU--------------------- (((((((..((((((((((......))))))))))..))))).))....(..(((((...........--....)))))..)................--------------------- ( -18.96, z-score = -2.62, R) >droVir3.scaffold_12855 1104153 79 + 10161210 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCACCAAUUGAUUUUCACUACGG-CAUUUGCUCGCACUUUCAUCCCAAGUUUC--------------------------------------- (((((((..((((..(((((....)))))..))))..))))).))..(((-(....)).))...................--------------------------------------- ( -17.00, z-score = -0.98, R) >droMoj3.scaffold_6540 9973595 118 + 34148556 GUAAAAUUGAUUGAAGCGUUUGUUGGCGCGCCAAUUGAUUUGCACUAUUG-CAUUUGCCCGCACUUACAACCUAAAAUCUCUACGUAGCCUCUAGCUUCGAGUUGGCCUGGCCUCUGUC (((((((..((((..(((((....)))))..))))..)))((((....))-))...........))))................(..(((.(((((.....)))))...)))..).... ( -25.30, z-score = 0.43, R) >consensus GUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAG_UACGUACACGAA____UAUGUUGCAGCACCCCUCCCGACUCCCAUU______________________ (.(((((..(((((((((((....)))))))))))..))))))............................................................................ (-18.37 = -18.78 + 0.41)

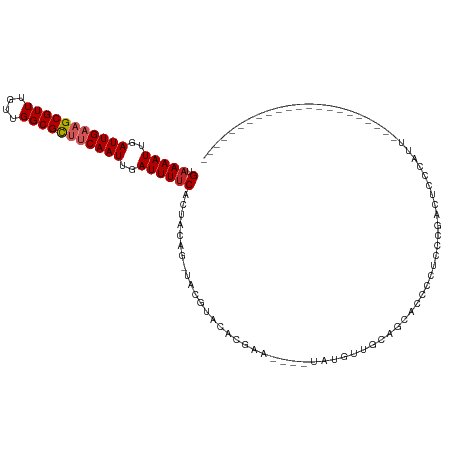
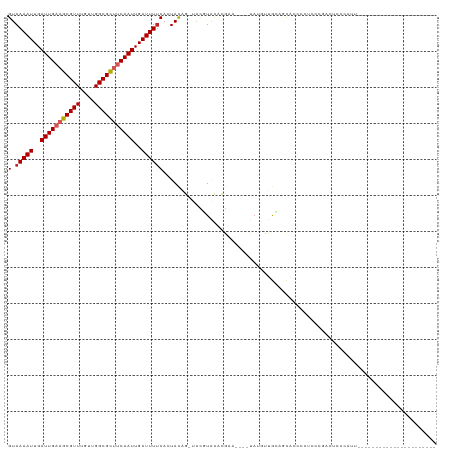
| Location | 2,420,433 – 2,420,549 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Shannon entropy | 0.44743 |
| G+C content | 0.38207 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.24 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2420433 116 - 27905053 -UUGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUACGAAUACGUAUGUUGCAGCACC--- -.(((((..((.(((((((((..-..--.....)).)))))))((((..(((((((((((....)))))))))))..))))........((((((((....))))))))))..)))))..--- ( -39.70, z-score = -4.41, R) >droSim1.chr3R 2448257 116 - 27517382 -UUGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUACGAAUACGUAUGUUGCAGCACC--- -.(((((..((.(((((((((..-..--.....)).)))))))((((..(((((((((((....)))))))))))..))))........((((((((....))))))))))..)))))..--- ( -39.70, z-score = -4.41, R) >droSec1.super_6 2514465 116 - 4358794 -UUGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUACGAAUACGUAUGUUGCAGCACC--- -.(((((..((.(((((((((..-..--.....)).)))))))((((..(((((((((((....)))))))))))..))))........((((((((....))))))))))..)))))..--- ( -39.70, z-score = -4.41, R) >droYak2.chr3R 18336753 110 - 28832112 -CUGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUAC------GUAUGUUGCAGCACC--- -.(((((..((.(((((((((..-..--.....)).)))))))((((..(((((((((((....)))))))))))..))))........(((((....)------))))))..)))))..--- ( -36.40, z-score = -3.81, R) >droEre2.scaffold_4770 2663121 110 - 17746568 -GUGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCAGUACAGUACGUAUAC------GUAUGUUGCAGCACC--- -((((((..((.(((((((((..-..--.....)).)))))))((((..(((((((((((....)))))))))))..))))........(((((....)------))))))..)))))).--- ( -39.30, z-score = -4.61, R) >droAna3.scaffold_13340 13645502 110 - 23697760 -AUGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGUUCCAAUUGAUUUUCACUACAGUACGUAUCC------GUGUGUUGCACCGCC--- -..((.(((.........((((.-(.--(((..(((...((((((((..((((.((((((....)))))).))))..))))).)))...)))..)))..------).))))..))).)).--- ( -25.30, z-score = -0.76, R) >dp4.chr2 10835685 102 + 30794189 -AGGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAGUCCCUACCG-----UAUAUGC------------ -.(((((((.(((((((((((..-..--.....)).)))))))))((..(((((((((((....)))))))))))..))......))))))).......-----.......------------ ( -30.70, z-score = -3.65, R) >droPer1.super_3 4671192 111 + 7375914 -AGGCUGUGAUUUAUUUUGCACU-CA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAGUCCCUACCG-----UAUAUGCUGUACGGCU--- -.(((((((.(((((((((((..-..--.....)).)))))))))((..(((((((((((....)))))))))))..))......)))))))....(((-----(((.....))))))..--- ( -37.10, z-score = -4.34, R) >droWil1.scaffold_181089 647023 117 - 12369635 -UUACUGUGAUUUAUUUUGCACUCCA--AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGCCGGCGUUUCAAUUGAUUUUCACUACAGUCAGUUUCC---UUCACAUCCCAUAGAACUACC -..((((((.(((((((((((..(..--..)..)).)))))))))((..((((((((((......))))))))))..))......)))))).(((((..---.............)))))... ( -25.66, z-score = -2.51, R) >droVir3.scaffold_12855 1104157 112 + 10161210 ACUGAUGUGAUUUAUUUUGCACU-CA--CUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCACCAAUUGAUUUUCACUACGGCAUUUGCUCGCACUUUCAUCCCAAG-------- ..(((((..(......)..)).)-))--..((.((((..((((((((..((((..(((((....)))))..))))..))))).)))..))))...))..................-------- ( -26.50, z-score = -1.36, R) >droMoj3.scaffold_6540 9973634 116 + 34148556 ACUGAUGUGAUUUAUUUUGCACU-CA--CUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCGCCAAUUGAUUUGCACUAUUGCAUUUGCCCGCACUUACAACCUAAAAUCU---- ..(((((..(......)..)).)-))--.(((.(((...(((.((((..((((..(((((....)))))..))))..))))..)))...((....))..)))...)))...........---- ( -23.20, z-score = -0.21, R) >droGri2.scaffold_14830 4704901 96 + 6267026 ----AUGUGAUUUAUUUUGCACU-CAGUGUGUUUGCUAAAGUAAAAUUGAUUGAAGCGUUUGUUGGUGCAUCAAUUGAUUUCCUCUAU-GCCUUAACACCCC--------------------- ----.((..(......)..))..-..((((....((...((..((((..(((((.(((........))).)))))..))))...))..-))....))))...--------------------- ( -19.30, z-score = -1.26, R) >consensus _UUGCUGUGAUUUAUUUUGCACU_CA__AUGUUUGCCAAAGUAAAAUUGAUUGAAGCGUUUGUUGGCGCUUCAAUUGAUUUUCACUACAGUACGUAUAC______GUAUGUUGCAGCACC___ .....((((.(((((((((((............))).))))))))((..((((..(((((....)))))..))))..))......)))).................................. (-15.49 = -15.24 + -0.25)
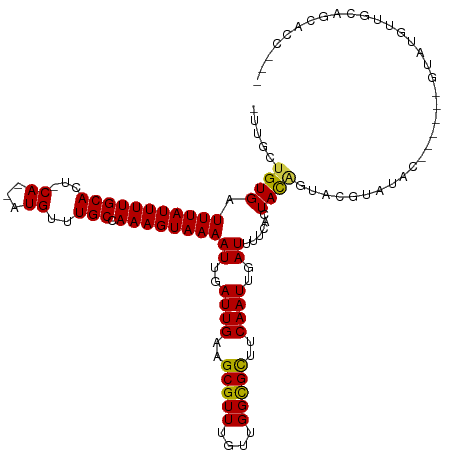
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:57 2011