| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,382,175 – 2,382,267 |
| Length | 92 |
| Max. P | 0.857370 |

| Location | 2,382,175 – 2,382,267 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Shannon entropy | 0.37573 |
| G+C content | 0.47330 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.55 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
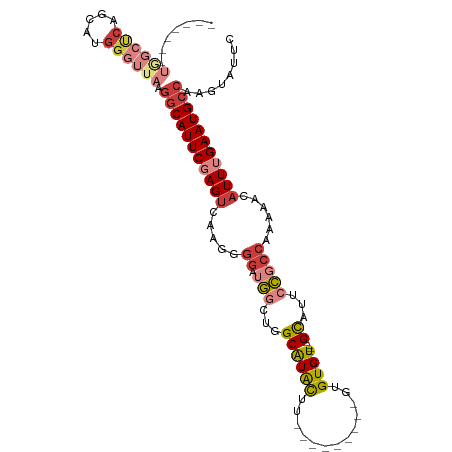
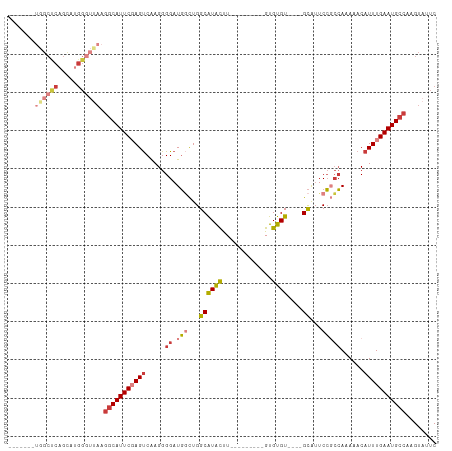
>dm3.chr3R 2382175 92 + 27905053 -------UGGCUCAGCAUGGGUAAAGGCAUUCGAGUCAAGGGGAUGGCUGGCAUACUU---------GUGUGU----GCAUUCCGCCAAAAACAUUUGAAUGCCAAGUAUUC -------..((((.....))))...(((((((((((.....((.(((...((((((..---------..))))----))...)))))......)))))))))))........ ( -32.30, z-score = -2.44, R) >droSim1.chr3R 2410103 92 + 27517382 -------UGGCUCAGCAUGGGUUAAGGCAUUCGAGUCAAGGGGAUGGCUGGCAUACUU---------GUGUGU----GCAUUCCGCCAAAAACAUUUGAAUGCCAAGUAUUC -------((((((.....)))))).(((((((((((.....((.(((...((((((..---------..))))----))...)))))......)))))))))))........ ( -33.50, z-score = -2.76, R) >droSec1.super_6 2475608 92 + 4358794 -------UGGCUCAGCAUGGGUUAAGGCAUUCGAGUCAAGGGGAUGGCUGGCAUACUU---------GUGUGU----GCAUUCCGCCAAAAACAUUUGAAUGCCAAGUAUUC -------((((((.....)))))).(((((((((((.....((.(((...((((((..---------..))))----))...)))))......)))))))))))........ ( -33.50, z-score = -2.76, R) >droYak2.chr3R 18297283 92 + 28832112 -------UGGCUCAGUAAGGGUUAAGGCAUUCCAGUCAAGGGGAUGGCUGGCAUACUU---------GUGUGU----GCAUUCCGCCAAAUACAUUUGAAUGCCAAGUAUUC -------((((((.....)))))).(((((((((((((......))))))((((((..---------..))))----))..................)))))))........ ( -30.10, z-score = -1.83, R) >droEre2.scaffold_4770 2623733 92 + 17746568 -------UGGCUCAGUAUGGGUUAAGGCAUUCGAGUCAAGGGGAUGGCUGGCAUACUU---------GUGUGU----GCAUUCCGCCAAAAACAUUUGAAUGCCAAGUAUUC -------((((((.....)))))).(((((((((((.....((.(((...((((((..---------..))))----))...)))))......)))))))))))........ ( -33.50, z-score = -3.15, R) >droAna3.scaffold_13340 13607320 102 + 23697760 UGGCCCAGAGCCCAGCAUGGAAUAAGGCAUUCGAGCCGAGGGG-UGUCUGGCGUGCCU---------GUGUGUAAGUGCAUUCCUCCAAAAACAUUUGAAUGCCAAGUAUUC .(((.....))).......(((((.((((((((((..((((((-(((((.((..(...---------.)..)).)).)))))))))........))))))))))...))))) ( -29.60, z-score = -0.12, R) >droPer1.super_3 4632492 92 - 7375914 --------------------GGGCGCACAUUCUAGUCAAGGGGAUUUUUGGCAUAUUUCCUCUGUGUGUGUGUUUGUGUGUGCACGAAAAAACAUUUGAAUGCCAAGUAUUC --------------------.(((((((((........((((((.............))))))))))))((.((((((....))))))...))........)))........ ( -22.82, z-score = -0.47, R) >consensus _______UGGCUCAGCAUGGGUUAAGGCAUUCGAGUCAAGGGGAUGGCUGGCAUACUU_________GUGUGU____GCAUUCCGCCAAAAACAUUUGAAUGCCAAGUAUUC .......((((((.....)))))).(((((((((((....((((((....((((((...........)))))).....)))))).........)))))))))))........ (-18.72 = -20.55 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:53 2011