| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,373,327 – 2,373,426 |
| Length | 99 |
| Max. P | 0.518854 |

| Location | 2,373,327 – 2,373,426 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.71 |
| Shannon entropy | 0.51580 |
| G+C content | 0.40812 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -8.41 |
| Energy contribution | -8.71 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
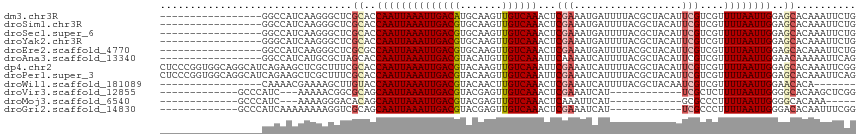
>dm3.chr3R 2373327 99 - 27905053 -----------------GGCCAUCAAGGGCUCGCACCAAUUAAAUUGACAUGCAAGUUGUCAAACUCGAAAUGAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCUG -----------------((((......)))).((.(((((((((.(((((((...((.((((.........))))...))....)))..)))).))))))))).)).......... ( -22.70, z-score = -1.63, R) >droSim1.chr3R 2401287 99 - 27517382 -----------------GGCCAUCAAGGGCUCGCACCAAUUAAAUUGACGUGCAAGUUGUCAAACUCGAAAUGAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCUG -----------------((((......)))).((.(((((((((.(((((.((.....((((.........)))).....))......))))).))))))))).)).......... ( -24.90, z-score = -2.01, R) >droSec1.super_6 2466754 99 - 4358794 -----------------GGCCAUCAAGGGCUCGCACCAAUUAAAUUGACGUGCAAGUUGUCAAACUCGAAAUGAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCUG -----------------((((......)))).((.(((((((((.(((((.((.....((((.........)))).....))......))))).))))))))).)).......... ( -24.90, z-score = -2.01, R) >droYak2.chr3R 18288156 99 - 28832112 -----------------GGGCAUCAAGGGCUCGCACCAAUUAAAUUGACGUGCAAGUUGUCAAACUCGAAAUGAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCUG -----------------((((.......))))((.(((((((((.(((((.((.....((((.........)))).....))......))))).))))))))).)).......... ( -24.70, z-score = -1.87, R) >droEre2.scaffold_4770 2614288 99 - 17746568 -----------------GGCCAUCAAGGGCUCGCGCCAAUUAAAUUGACGUGCAAGUUGUCAAACUCGAAAUGAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCUG -----------------((((......)))).((.(((((((((.(((((.((.....((((.........)))).....))......))))).))))))))).)).......... ( -25.40, z-score = -1.72, R) >droAna3.scaffold_13340 13594746 99 - 23697760 -----------------GGCCAUCAUGCGCUAGCACCAAUUAAAUUGACGUACAUGUUGUCAAAUUCAAAAUCAUUUUACGCUACAUUCGUCGUUUUAAUUGGAACAAAAAUUCAG -----------------........(((....)))(((((((((.(((((...(((((((.((((........)))).)))..)))).))))).)))))))))............. ( -20.30, z-score = -2.46, R) >dp4.chr2 10788337 116 + 30794189 CUCCCGGUGGCAGGCAUCAGAAGCUCGCUUUCGCACCAAUUAAAUUGACGUACAAGUUGUCAAAUUCGAAAUCAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCGG ...((((.(((..((.......))..)))...((.(((((((((.(((((....(((.((.((((........)))).))))).....))))).))))))))).))......)))) ( -28.90, z-score = -2.08, R) >droPer1.super_3 4623643 116 + 7375914 CUCCCGGUGGCAGGCAUCAGAAGCUCGCUUUCGCACCAAUUAAAUUGACGUACAAGUUGUCAAAUUCGAAAUCAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCAG .....((.(((..((.......))..))).))((.(((((((((.(((((....(((.((.((((........)))).))))).....))))).))))))))).)).......... ( -27.70, z-score = -2.10, R) >droWil1.scaffold_181089 589967 92 - 12369635 -----------------CAAAACGAAAAGCUUGUACCAAUUAAAUUGACGUACAACUUGUCAAACUCGAAAUCAUUUUACGCUACAAUCGUCGUUUUAAUUGGAACACA------- -----------------..............(((.(((((((((.(((((.......(((.(((..........))).))).......))))).))))))))).)))..------- ( -15.94, z-score = -1.48, R) >droVir3.scaffold_12855 1042590 88 + 10161210 -------------GCCCAUC---AAAAACGGCGCAGCAAUUAAAUUGACGUACGAGUUGUCAAACUCGAAAUCAU------------UCGCUCUUUUAAUUGGGGCACAAGCUCGG -------------((((...---.((((.(((((..((((...))))..)).((((((....)))))).......------------..))).)))).....)))).......... ( -20.90, z-score = -1.45, R) >droMoj3.scaffold_6540 9920004 83 + 34148556 -------------GCCCAUC---AAAAGGGACACAGCAAUUAAAUUGACGUACGAGUUGUCAAACUCAAAUUCAU------------GCGCCCUUUUAAUUGGGGCACAAA----- -------------((((...---(((((((.(.(((((((...)))).)....(((((....))))).......)------------).)))))))).....)))).....----- ( -23.10, z-score = -2.85, R) >droGri2.scaffold_14830 4651788 91 + 6267026 -------------GCCCAUCAAAAAAAAGGUCGCAGCAAUUAAAUUGACGUACGAGUUGUCAAACUCGAAAUCAU------------UCGCCCUUUUAAUUGGGACACAAUUUCGG -------------.((((......((((((.(((..((((...))))..)).((((((....)))))).......------------..).))))))...))))............ ( -18.30, z-score = -1.21, R) >consensus _________________GGCCAUCAAGGGCUCGCACCAAUUAAAUUGACGUACAAGUUGUCAAACUCGAAAUCAUUUUACGCUACAUUCGUCGUUUUAAUUGGAGCACAAAUUCUG ................................((..((((((((((((((.......))))))...(((..................)))....))))))))..)).......... ( -8.41 = -8.71 + 0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:52 2011