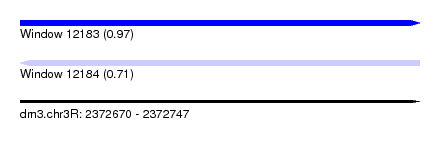
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,372,670 – 2,372,747 |
| Length | 77 |
| Max. P | 0.974861 |

| Location | 2,372,670 – 2,372,747 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 71.47 |
| Shannon entropy | 0.56667 |
| G+C content | 0.46745 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.71 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
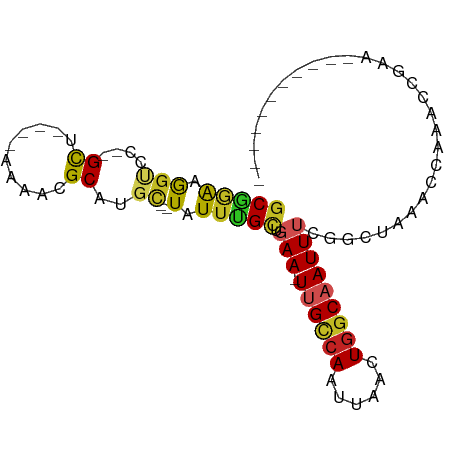
>dm3.chr3R 2372670 77 + 27905053 GCGAAAGGUCC--GCU----AAAACGCAUGC--UAUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUUGGCUGAUCCAAAACGAACC--------- ......((((.--(((----(....(((...--....))).(((-((((((......))))))))))))).))))...........--------- ( -20.50, z-score = -2.36, R) >droSim1.chr3R 2400587 77 + 27517382 GCGGAAGGUCC--GCU----AAAACGCAUGC--UAUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUCGGCUGAACCAAACCGAACC--------- .(((..(((.(--((.----.....))....--.....((((((-((((((......)))))).)))))).).)))...)))....--------- ( -23.20, z-score = -2.80, R) >droSec1.super_6 2466104 77 + 4358794 GCGGAAGGUCC--GCU----AAAACGCAUGC--UAUUUGCUGAA-UUGUCAAUUAACUGGCAAUUUCGGCUGGACCAAAUUGAACC--------- ......(((((--((.----.....))....--.....((((((-((((((......)))))).)))))).)))))..........--------- ( -24.50, z-score = -3.09, R) >droYak2.chr3R 18287553 82 + 28832112 GCGGAAGGUCC--GCC----AAAACGCAUGC--UAUUCGCUGAA-UUGCCAAUUAACUGGCAAUUUCGGCUAAACCUAACCGAACCGAACC---- .(((.((((..--((.----.....))....--.....((((((-((((((......)))))).))))))...))))..))).........---- ( -23.50, z-score = -2.69, R) >droEre2.scaffold_4770 2613523 85 + 17746568 GCGGAAGGUCC--GCU----AAAACGCAUGC--UAUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUCGGCUGAACCAU-CCGAACCAUCCGAACC .((((.(((.(--((.----.....))....--.....((((((-((((((......)))))).))))))........-..).))).)))).... ( -27.10, z-score = -3.43, R) >droAna3.scaffold_13340 13594222 70 + 23697760 GCGGAAGGUCC--GCU----AAAACGCAUGC--UAUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUCGACCAAAAACGA---------------- ((((.....))--)).----.....(((...--....))).(((-((((((......))))))))).............---------------- ( -19.00, z-score = -2.23, R) >dp4.chr2 10787442 78 - 30794189 GCGGAAGGUCCAGGUCCGCUAAAACGCGUGC--UAUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUCGGCUAAAGCCAACC-------------- ..((..(((...((..(((......)))..)--)....((((((-((((((......)))))).))))))....)))..))-------------- ( -26.10, z-score = -2.22, R) >droPer1.super_3 4622726 78 - 7375914 GCGGAAGGUCCAGGUCCGCUAAAACGCGUGC--UAUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUCGGCUAAAGCCAACC-------------- ..((..(((...((..(((......)))..)--)....((((((-((((((......)))))).))))))....)))..))-------------- ( -26.10, z-score = -2.22, R) >droWil1.scaffold_181089 588929 83 + 12369635 GCAGAAGGUCC--GCCC---AGAACGCAUUCGGUUUUUGCUGAA-UUGCCAAUUAACUGGCAAUUUCCACUAAAACGUACAAACGAAUC------ (((((((..((--(...---..........)))))))))).(((-((((((......))))))))).........(((....)))....------ ( -17.32, z-score = -0.80, R) >droMoj3.scaffold_6540 9919009 73 - 34148556 GCACACAGCCA--GCU-------ACGCAUGU--UAUUUGUCGAAAUCGCCAAUUAACUGGCAAUUGCUCCAAAUGGAAUGUCUC----------- (((....((((--((.-------..))..((--((.(((.((....)).))).))))))))...)))(((....))).......----------- ( -15.70, z-score = -0.68, R) >droVir3.scaffold_12855 1041221 72 - 10161210 UCUGGGAAGCU--GCU-------ACGCAUGA--UAUUUGCUGAA-UCGACAAUUAACUAGCAAUUGCGCCAUGUGAAAGACUCU----------- ....(((..((--...-------.((((((.--...((((((..-............))))))......))))))..))..)))----------- ( -12.14, z-score = 0.60, R) >consensus GCGGAAGGUCC__GCU____AAAACGCAUGC__UAUUUGCUGAA_UUGCCAAUUAACUGGCAAUUUCGGCUAAACCAAACCGAA___________ ......((((...............(((.........))).(((.((((((......)))))).)))))))........................ (-10.23 = -10.71 + 0.48)

| Location | 2,372,670 – 2,372,747 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 71.47 |
| Shannon entropy | 0.56667 |
| G+C content | 0.46745 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -9.96 |
| Energy contribution | -10.25 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.714185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
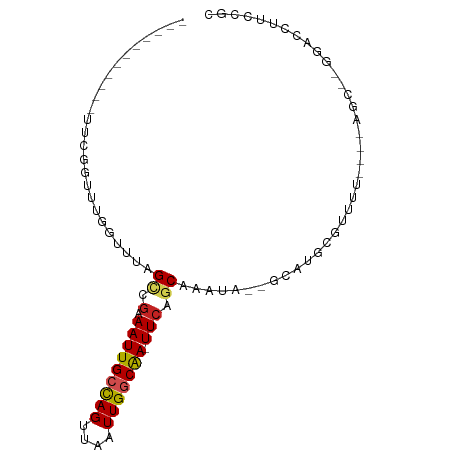
>dm3.chr3R 2372670 77 - 27905053 ---------GGUUCGUUUUGGAUCAGCCAAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAUA--GCAUGCGUUUU----AGC--GGACCUUUCGC ---------((((((((((((.....))))(((((((((....)))))))-))..(((....--...))).....----)))--)))))...... ( -23.10, z-score = -2.39, R) >droSim1.chr3R 2400587 77 - 27517382 ---------GGUUCGGUUUGGUUCAGCCGAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAUA--GCAUGCGUUUU----AGC--GGACCUUCCGC ---------....(((...(((((.((.(((.(((((((....)))))))-))).)).....--....((.....----.))--)))))..))). ( -25.50, z-score = -2.51, R) >droSec1.super_6 2466104 77 - 4358794 ---------GGUUCAAUUUGGUCCAGCCGAAAUUGCCAGUUAAUUGACAA-UUCAGCAAAUA--GCAUGCGUUUU----AGC--GGACCUUCCGC ---------..........(((((.((..((((.((..((((.(((.(..-....)))).))--))..)))))).----.))--)))))...... ( -19.00, z-score = -1.27, R) >droYak2.chr3R 18287553 82 - 28832112 ----GGUUCGGUUCGGUUAGGUUUAGCCGAAAUUGCCAGUUAAUUGGCAA-UUCAGCGAAUA--GCAUGCGUUUU----GGC--GGACCUUCCGC ----.........(((..((((((.((((((((((((((....)))))).-....((.....--))....)))))----)))--)))))).))). ( -27.70, z-score = -2.01, R) >droEre2.scaffold_4770 2613523 85 - 17746568 GGUUCGGAUGGUUCGG-AUGGUUCAGCCGAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAUA--GCAUGCGUUUU----AGC--GGACCUUCCGC ....((((.(((((..-(((.(...((.(((.(((((((....)))))))-))).))....)--.)))((.....----.))--))))).)))). ( -27.90, z-score = -1.87, R) >droAna3.scaffold_13340 13594222 70 - 23697760 ----------------UCGUUUUUGGUCGAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAUA--GCAUGCGUUUU----AGC--GGACCUUCCGC ----------------........(((((((.(((((((....)))))))-))).((.....--)).(((.....----.))--)))))...... ( -18.90, z-score = -1.59, R) >dp4.chr2 10787442 78 + 30794189 --------------GGUUGGCUUUAGCCGAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAUA--GCACGCGUUUUAGCGGACCUGGACCUUCCGC --------------((((.(((..(((((.(((((((((....)))))))-))..((.....--)).)).)))..))).)))).((.....)).. ( -27.10, z-score = -2.35, R) >droPer1.super_3 4622726 78 + 7375914 --------------GGUUGGCUUUAGCCGAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAUA--GCACGCGUUUUAGCGGACCUGGACCUUCCGC --------------((((.(((..(((((.(((((((((....)))))))-))..((.....--)).)).)))..))).)))).((.....)).. ( -27.10, z-score = -2.35, R) >droWil1.scaffold_181089 588929 83 - 12369635 ------GAUUCGUUUGUACGUUUUAGUGGAAAUUGCCAGUUAAUUGGCAA-UUCAGCAAAAACCGAAUGCGUUCU---GGGC--GGACCUUCUGC ------((..((((((...(((((.((.(((.(((((((....)))))))-))).)).)))))))))))...)).---..((--((.....)))) ( -20.80, z-score = -0.60, R) >droMoj3.scaffold_6540 9919009 73 + 34148556 -----------GAGACAUUCCAUUUGGAGCAAUUGCCAGUUAAUUGGCGAUUUCGACAAAUA--ACAUGCGU-------AGC--UGGCUGUGUGC -----------...((((.(((((((..(.(((((((((....))))))))).)..))))).--....((..-------.))--.))..)))).. ( -18.40, z-score = -0.47, R) >droVir3.scaffold_12855 1041221 72 + 10161210 -----------AGAGUCUUUCACAUGGCGCAAUUGCUAGUUAAUUGUCGA-UUCAGCAAAUA--UCAUGCGU-------AGC--AGCUUCCCAGA -----------...(((........)))((..(((((((((.......))-)).)))))...--...(((..-------.))--)))........ ( -9.20, z-score = 1.98, R) >consensus ___________UUCGGUUUGGUUUAGCCGAAAUUGCCAGUUAAUUGGCAA_UUCAGCAAAUA__GCAUGCGUUUU____AGC__GGACCUUCCGC .........................((.(((.(((((((....))))))).))).))...................................... ( -9.96 = -10.25 + 0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:51 2011