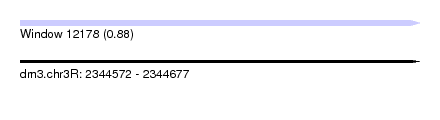
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,344,572 – 2,344,677 |
| Length | 105 |
| Max. P | 0.884806 |

| Location | 2,344,572 – 2,344,677 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.73165 |
| G+C content | 0.39352 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -6.84 |
| Energy contribution | -6.58 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
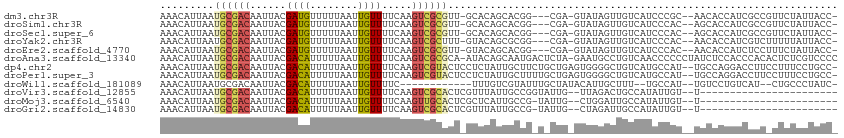
>dm3.chr3R 2344572 105 + 27905053 AAACAUUAAUGCGACAAUUACGAUGUUUUUAAUUGUUUUCAAGUCGCGUU-GCACAGCACGG---CGA-GUAUAGUUGUCAUCCCGC--AACACCAUCGCCGUUCUAUUACC- ......(((((((((....(((((.......)))))......))))))))-)...((.((((---(((-.....(((((......))--)))....))))))).))......- ( -30.80, z-score = -3.86, R) >droSim1.chr3R 2371238 105 + 27517382 AAACAUUAAUGCGACAAUUACGAUGUUUUUAAUUGUUUUCAAGUCGCGUU-GCACAGCACGG---CGA-GUAUAGUUGUCAUCCCAC--AGCACCAUCGCCGUUCUAUUACC- ......(((((((((....(((((.......)))))......))))))))-)...((.((((---(((-.....(((((......))--)))....))))))).))......- ( -30.50, z-score = -4.05, R) >droSec1.super_6 2429084 105 + 4358794 AAACAUUAAUGCGACAAUUACGAUGUUUUUAAUUGUUUUCAAGUCGCGUU-GCACAGCACGG---CGA-GUAUAGUUGUCAUCCCAC--AGCACCAUCGCCGUUCUAUUACC- ......(((((((((....(((((.......)))))......))))))))-)...((.((((---(((-.....(((((......))--)))....))))))).))......- ( -30.50, z-score = -4.05, R) >droYak2.chr3R 18259180 105 + 28832112 AAACAUUAAUGCGACAAUUACGAUGUUUUUAAUUGUUUUCAAGUCGCUUU-GUACAGCGCGG---CGA-GUAUAGUUGUCAUCCCAC--AACACCAUCGUCUUUUUAUUACC- ..(((.....(((((....(((((.......)))))......)))))..)-)).......((---(((-.....(((((......))--)))....)))))...........- ( -20.10, z-score = -1.19, R) >droEre2.scaffold_4770 2585678 105 + 17746568 AAACAUUAAUGCGACAAUUACGAUGUUUUUAAUUGUUUUCAAGUCGCGUU-GUACAGCACGG---CGA-GUAUAGUUGUCAUCCCAC--AACACCAUCUCCUUUCUAUUACC- ......(((((((((....(((((.......)))))......))))))))-)........((---.((-.....(((((......))--)))....)).))...........- ( -20.20, z-score = -1.66, R) >droAna3.scaffold_13340 13565262 111 + 23697760 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUCGCGCA-AUACAGCAAUGACUCUA-GAAUGCCUGUCAACCCCCCUAUCUCCACCCACACUCUCGUCCCC ..........(((((......((((........)))).....)))))((.-.....))..((((....-........))))................................ ( -13.50, z-score = -1.32, R) >dp4.chr2 10759156 110 - 30794189 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUCGUACUCCUCUAUUGCUUCUGCUGAGUGGGGCUGUCAUGCCAU--UGCCAGGACCUUCCUUUCCUGCC- .........((((((......((((........)))).....))))))(..(((....((....)).)))..)(((......)))..--...(((((........)))))..- ( -24.10, z-score = -0.98, R) >droPer1.super_3 4593541 110 - 7375914 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUCGUACUCCUCUAUUGCUUUUGCUGAGUGGGGCUGUCAUGCCAU--UGCCAGGACCUUCCUUUCCUGCC- .........((((((......((((........)))).....))))))(..(((....((....)).)))..)(((......)))..--...(((((........)))))..- ( -24.10, z-score = -1.00, R) >droWil1.scaffold_181089 11803708 94 - 12369635 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUC------------UUUGUCGUAUUUGCUAUACAUUGCUUU--UGCCAU--UGUCCUGUCAU--CUGCCCUAUC- .......((((((((((....((((........))))...------------.)))))))))).((...(((..((...--.))...--)))...))...--..........- ( -15.50, z-score = -2.27, R) >droVir3.scaffold_12855 1008367 86 - 10161210 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUCGCACUCGUUUAUUGCCGGUAUUG--UUAGACUGCCAUAUUGU--U----------------------- .((((.((.((((((......((((........)))).....)))))).............((((...--......)))).)).)))--)----------------------- ( -13.90, z-score = -0.18, R) >droMoj3.scaffold_6540 9886513 85 - 34148556 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUUGCACUCGCUCAUUGCCG-UAUUG--CUGGAUUGCCAUAUUGU--U----------------------- ...((.((((((((((((((.........)))))))...(((...((....))...))).))-)))))--.))..............--.----------------------- ( -12.10, z-score = 0.86, R) >droGri2.scaffold_14830 4621486 85 - 6267026 AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUCGCACUCGUUUAUUGCCG-UAUUG--CUAGAUUGCCAUAUUGU--U----------------------- .........((((((......((((........)))).....))))))..........((.(-(((.(--(......)).)))).))--.----------------------- ( -14.00, z-score = -0.50, R) >consensus AAACAUUAAUGCGACAAUUACGACAUUUUUAAUUGUUUUCAAGUCGCACU_GUACAGCGCGG_U_CUA_GUAUGGUUGUCAUCCCAC__AGCACCAUCGCC_UUCUAUUACC_ .........((((((......((((........)))).....))))))................................................................. ( -6.84 = -6.58 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:46 2011