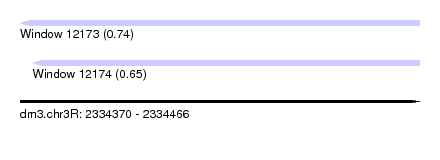
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,334,370 – 2,334,466 |
| Length | 96 |
| Max. P | 0.741268 |

| Location | 2,334,370 – 2,334,466 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Shannon entropy | 0.35517 |
| G+C content | 0.41990 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
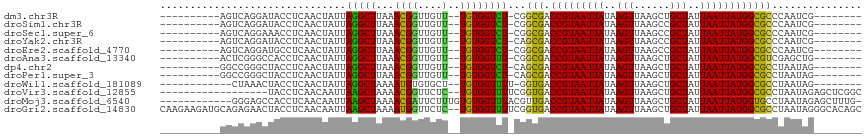
>dm3.chr3R 2334370 96 - 27905053 ----------AGUCAGGAUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCCAAUCG-------- ----------......((.(((.((((..((.......))..))))..--...))).)-)((((.(((((((((..(((......)))..)))))))))))))......-------- ( -25.60, z-score = -1.43, R) >droSim1.chr3R 2366077 96 - 27517382 ----------AGUCAGGAUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAAUCG-------- ----------......((.(((.((((..((.......))..))))..--...))).)-)((((.(((((((((..(((......)))..)))))))))))))......-------- ( -24.90, z-score = -1.05, R) >droSec1.super_6 2423594 96 - 4358794 ----------AGUCAGGAAACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAAUCG-------- ----------......((.(((.((((..((.......))..))))..--...))).)-)((((.(((((((((..(((......)))..)))))))))))))......-------- ( -24.90, z-score = -1.14, R) >droYak2.chr3R 18254216 96 - 28832112 ----------AGUCAGGAUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CAGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAAUCG-------- ----------.....((...............((((((((((((((((--.(......-)))))))))).........)))))))(((((.....)))))..)).....-------- ( -25.10, z-score = -1.57, R) >droEre2.scaffold_4770 2580284 96 - 17746568 ----------AGUCAGGAUGCCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAAUCG-------- ----------.........((((........)))).....((((((..--.(......-)((((.(((((((((..(((......)))..)))))))))))))))))))-------- ( -25.30, z-score = -0.84, R) >droAna3.scaffold_13340 13561472 96 - 23697760 ----------ACUCGGGCCACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUUU-CGGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGUCGAGCUG-------- ----------...((((((((..((((..((.......))..))))..--.)))))((-(((((.(((((((((..(((......)))..)))))))))))))))))))-------- ( -31.10, z-score = -2.53, R) >dp4.chr2 10751723 96 + 30794189 ----------GGCCGGGCUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CAGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAAUAG-------- ----------....((....))....(((((((((....(((((((((--.(......-))))))))))................(((((.....))))))))))))))-------- ( -29.80, z-score = -1.96, R) >droPer1.super_3 4585972 96 + 7375914 ----------GGCCGGGCUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CAGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAAUAG-------- ----------....((....))....(((((((((....(((((((((--.(......-))))))))))................(((((.....))))))))))))))-------- ( -29.80, z-score = -1.96, R) >droWil1.scaffold_181089 557072 94 - 12369635 ------------CUAAACUACCUCAACUAUUAGGCUAAAAUGUGUGCU--UGUGGUUUU-GGUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAAUAG-------- ------------..............(((((((((......((..(((--(((((((..-(....)...))))))))))...)).(((((.....))))))))))))))-------- ( -28.40, z-score = -3.58, R) >droVir3.scaffold_12855 1001474 97 + 10161210 ------------------UACCUCAACAAUUAAGCUAAAACGGUUCUC--UGUGGUUUUCGGUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAAUAGAGCUCGGC ------------------...........(..((((...((((....)--))).......((((.(((((((((..(((......)))..)))))))))))))......))))..). ( -23.70, z-score = -1.20, R) >droMoj3.scaffold_6540 9880030 104 + 34148556 ------------GGGAGCCACCUCAACAAUUAAGCUAAAACGAUUCUUUGUGUGGUUUACGUUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGUGCCUAAUAGAGCUUUG- ------------.((((((....((((...(((((((..((((....)))).))))))).))))((((((((((..(((......)))..))))))))))........).))))).- ( -25.00, z-score = -1.44, R) >droGri2.scaffold_14830 4616221 115 + 6267026 CAAGAAGAUGCAGAGAACUACCUCAACAAUUAAGCUAAAAUGGUUCUC--UGUGGUUUUCGGUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAAUAGGGCACAGC ...(((((((((((((((((....................))))))))--))).))))))((((.(((((((((..(((......)))..))))))))))))).............. ( -34.25, z-score = -2.88, R) >consensus __________AGUCAGGCUACCUCAACUAUUAGGCUUAAACGGUUGUU__UGUGGUCU_CGGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAAUAG________ ...................(((.((.......((((.....)))).....)).))).....(((.(((((((((..(((......)))..))))))))))))............... (-17.03 = -16.73 + -0.30)


| Location | 2,334,373 – 2,334,466 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Shannon entropy | 0.33396 |
| G+C content | 0.41175 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
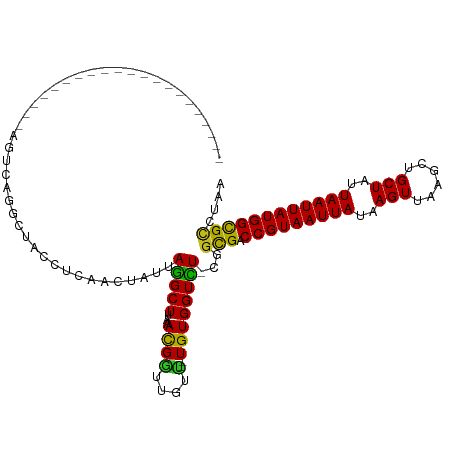
>dm3.chr3R 2334373 93 - 27905053 ---------------------AGUCAGGAUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCCAA ---------------------......((.(((.((((..((.......))..))))..--...))).)-)((((.(((((((((..(((......)))..)))))))))))))... ( -25.60, z-score = -1.44, R) >droSim1.chr3R 2366080 93 - 27517382 ---------------------AGUCAGGAUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAA ---------------------......((.(((.((((..((.......))..))))..--...))).)-)((((.(((((((((..(((......)))..)))))))))))))... ( -24.90, z-score = -1.07, R) >droSec1.super_6 2423597 93 - 4358794 ---------------------AGUCAGGAAACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAA ---------------------......((.(((.((((..((.......))..))))..--...))).)-)((((.(((((((((..(((......)))..)))))))))))))... ( -24.90, z-score = -1.15, R) >droYak2.chr3R 18254219 93 - 28832112 ---------------------AGUCAGGAUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CAGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAA ---------------------.....((...............((((((((((((((((--.(......-)))))))))).........)))))))(((((.....)))))..)).. ( -25.10, z-score = -1.58, R) >droEre2.scaffold_4770 2580287 93 - 17746568 ---------------------AGUCAGGAUGCCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CGGCGACCGUAAUUAUAAGUUAAGCCGCUAUUAAUUAUGGCGCCCAA ---------------------.....((.((((..........((((((((((((((((--........-.))))))))).........)))))))............)))).)).. ( -25.25, z-score = -0.86, R) >droAna3.scaffold_13340 13561475 93 - 23697760 ---------------------ACUCGGGCCACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUUU-CGGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGUCGAG ---------------------.....((((((..((((..((.......))..))))..--.))))))(-(((((.(((((((((..(((......)))..))))))))))))))). ( -30.80, z-score = -2.97, R) >dp4.chr2 10751726 93 + 30794189 ---------------------GGCCGGGCUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CAGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAA ---------------------....(((((((..((((..((.......))..))))..--.)))))))-..(((.(((((((((..(((......)))..)))))))))))).... ( -26.60, z-score = -1.00, R) >droPer1.super_3 4585975 93 + 7375914 ---------------------GGCCGGGCUACCUCAACUAUUAGGCUUAAACGGUUGUU--UGUGGUCU-CAGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAA ---------------------....(((((((..((((..((.......))..))))..--.)))))))-..(((.(((((((((..(((......)))..)))))))))))).... ( -26.60, z-score = -1.00, R) >droWil1.scaffold_181089 557075 91 - 12369635 -----------------------CUAAACUACCUCAACUAUUAGGCUAAAAUGUGUGCU--UGUGGUUU-UGGUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAA -----------------------............((((((.((((..........)))--))))))).-.((((.(((((((((..(((......)))..)))))))))))))... ( -23.20, z-score = -1.97, R) >droVir3.scaffold_12855 1001485 86 + 10161210 -----------------------------UACCUCAACAAUUAAGCUAAAACGGUUCUC--UGUGGUUUUCGGUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAA -----------------------------.............(((((...((((....)--))))))))..((((.(((((((((..(((......)))..)))))))))))))... ( -20.50, z-score = -1.84, R) >droMoj3.scaffold_6540 9880040 103 + 34148556 -------------AGUUGG-GAUGGGAGCCACCUCAACAAUUAAGCUAAAACGAUUCUUUGUGUGGUUUACGUUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGUGCCUAA -------------.(((((-(.(((...))).))))))...(((((((..((((....)))).))))))).....((((((((((..(((......)))..))))))))))...... ( -26.80, z-score = -1.54, R) >droGri2.scaffold_14830 4616232 115 + 6267026 GGCAAUAAUAACAAGAAGAUGCAGAGAACUACCUCAACAAUUAAGCUAAAAUGGUUCUC--UGUGGUUUUCGGUGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAA ..............(((((((((((((((((....................))))))))--))).))))))((((.(((((((((..(((......)))..)))))))))))))... ( -34.25, z-score = -3.79, R) >consensus _____________________AGUCAGGCUACCUCAACUAUUAGGCUUAAACGGUUGUU__UGUGGUCU_CGGCGACCGUAAUUAUAAGUUAAGCUGCUAUUAAUUAUGGCGCCUAA ..............................(((.((.......((((.....)))).....)).))).....(((.(((((((((..(((......)))..)))))))))))).... (-17.03 = -16.73 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:43 2011