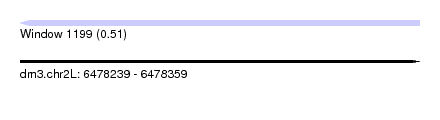
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,478,239 – 6,478,359 |
| Length | 120 |
| Max. P | 0.510205 |

| Location | 6,478,239 – 6,478,359 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Shannon entropy | 0.33568 |
| G+C content | 0.53889 |
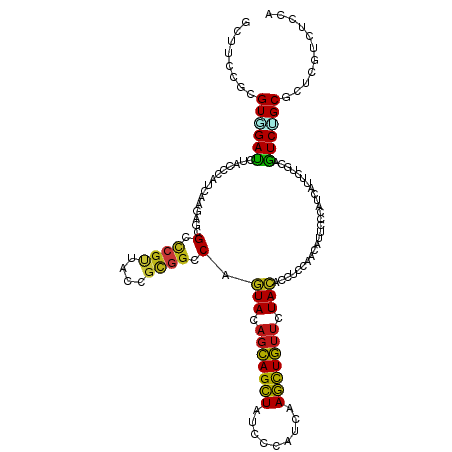
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.21 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
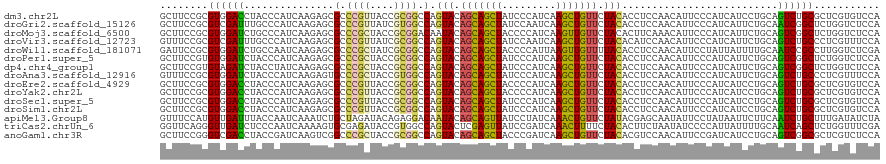
>dm3.chr2L 6478239 120 - 23011544 GCUUCCGCGUGGACCUACCCAUCAAGAGCGCCCGUUACCGCGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUCCUGCAGUCUGCGCUCGUGUCCA .....(((((((......))))...((((((((((....))))...(((.(((((((.........))))))).)))..............................))))))))).... ( -30.80, z-score = -2.40, R) >droGri2.scaffold_15126 3438937 120 - 8399593 GCUUCCGCGUCGAUUUGCCCAUCAAGAGCGCCCGUUAUCGUGGCCAGUACAGCAGCUAUCCAAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUUCUGCAAUCGGCUCUGGUCUCCA ....(((.(((((((.((.........(.(((((....)).)))).(((.(((((((.........))))))).))).......................)))))))))..)))...... ( -27.90, z-score = -1.80, R) >droMoj3.scaffold_6500 24778725 120 + 32352404 GCUUCCGCGUGGAUCUGCCCAUCAAGAGCGCCCGCUACCGCGGACAAUACAGCAGCUACCCCAUCAAGUUGUUCUACACUUCAAACAUUCCCAUCAUUCUGCAGUCGGCUCUGGUCUCCA ((....))(..((((.(((.((..((((.(.((((....)))).).....(((((((.........))))))).......................))))...)).)))...))))..). ( -24.60, z-score = 0.42, R) >droVir3.scaffold_12723 4484886 120 - 5802038 GUUUCCGCGUCGAUUUGCCCAUCAAGAGCGCCCGUUAUCGCGGCCAGUACAGCAGCUAUCCAAUCAAGCUGUUCUACACAUCCAACAUUCCCAUCAUUCUGCAGUCUGCCCUCGUUUCCA ......(((..((((.((.........(.(((((....)).)))).(((.(((((((.........))))))).))).......................)))))))))........... ( -21.40, z-score = -1.05, R) >droWil1.scaffold_181071 584559 120 - 1211509 GAUUCCGCGUGGAUCUGCCAAUCAAGAGCGCCCGCUAUCGCGGCCAGUACAGCAGCUACCCCAUUAAGUUGUUUUACACCUCCAACAUUCCUAUUAUUUUGCAAUCCGCCUUGGUCUCGA ((..(((.((((((.(((...........(.((((....)))).).(((.(((((((.........))))))).))).......................)))))))))..)))..)).. ( -29.20, z-score = -1.64, R) >droPer1.super_5 3135446 120 + 6813705 GCUUCCGUGUGGAUCUACCCAUCAAGAGCGCCCGCUACCGCGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUUCUGCAGUCGGCUCUGGUCUCCA .........((((...(((.....((((((.((((....)))).).(((.(((((((.........))))))).)))..............................)))))))).)))) ( -28.60, z-score = -1.03, R) >dp4.chr4_group1 1528721 120 - 5278887 GCUUCCGUGUAGAUCUACCUAUCAAGAGCGCCCGCUACCGCGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUUCUGCAGUCGGCUCUGGUCUCCA (((..(.((((((................(.((((....)))).).(((.(((((((.........))))))).)))....................)))))))..)))........... ( -27.00, z-score = -1.16, R) >droAna3.scaffold_12916 2577140 120 + 16180835 GUUUCCGCGUGGAUCUACCCAUCAAGAGUGCCCGCUACCGUGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUUCUGCAGUCUGCCCUCGUUUCCA ......((((((......))))..(((.(((((((....))))...(((.(((((((.........))))))).))).......................))).)))))........... ( -25.80, z-score = -1.89, R) >droEre2.scaffold_4929 15387033 120 - 26641161 GCUUCCGCGUGGACCUACCCAUCAAGAGCGCCCGUUACCGCGGCCAGUACAGCAGCUACCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUCCUGCAGUCUGCGCUCGUGUCCA .....(((((((......))))...((((((((((....))))...(((.(((((((.........))))))).)))..............................))))))))).... ( -30.80, z-score = -2.34, R) >droYak2.chr2L 15885799 120 + 22324452 GCUUCCGCGUGGACCUACCCAUCAAGAGCGCCCGUUACCGCGGCCAGUACAGCAGCUACCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUCCUGCAGUCUGCGCUCGUGUCCA .....(((((((......))))...((((((((((....))))...(((.(((((((.........))))))).)))..............................))))))))).... ( -30.80, z-score = -2.34, R) >droSec1.super_5 4527504 120 - 5866729 GCUUCCGCGUGGACCUACCCAUCAAGAGCGCCCGUUACCGCGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUCCUGCAGUCUGCGCUCGUGUCCA .....(((((((......))))...((((((((((....))))...(((.(((((((.........))))))).)))..............................))))))))).... ( -30.80, z-score = -2.40, R) >droSim1.chr2L 6269055 120 - 22036055 GCUUCCGCGUGGACCUACCCAUCAAGAGCGCCCGUUACCGCGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUCCUGCAGUCUGCGCUCGUGUCCA .....(((((((......))))...((((((((((....))))...(((.(((((((.........))))))).)))..............................))))))))).... ( -30.80, z-score = -2.40, R) >apiMel3.Group8 4152330 120 + 11452794 GUUUCCAUGUUGAUUUACCAAUCAAAUCUGCUAGAUACAGAGGACAAUACAGCAGUUAUCCUAUCAAACUGUUCUAUACGAGCAAUAUUCCUAUAAUUCUUCAAUCUGCUUUGAUAUCUA .........((((((....))))))(((.((.((((...(((((......(((((((.........))))))).((((.(((.....))).))))..))))).))))))...)))..... ( -17.00, z-score = -0.03, R) >triCas2.chrUn_6 185544 120 + 1398711 GGUUCAGGGUUGAUCUCCCAAUCAAAAGUGCGAGAUACCGUGGCCAGUACUCGAGUUAUCCGAUCAAACUUUUCUACACUUCUAAUAUCCCCAUUAUUUUGCAAUCAGCUCUGGUUUCGA ((.(((((((((((..............(((((((((..((((.........((((.....((.........))...)))).........))))))))))))))))))))))))..)).. ( -22.60, z-score = 0.35, R) >anoGam1.chr3R 28549632 120 - 53272125 GCUUCCGGGUCGACCUACCGAUCAAGUCGGCCCGCUACCGCGGCCAGUACAGCAGCUACCCGAUCAAGCUGUUCUACACGUCCAACAUUCCGAUCAUCCUGCAGUCGGCGCUCGUCUCCA ((....((((((((...........))))))))(((((.((((...(((.(((((((.........))))))).))).((..........))......)))).)).)))))......... ( -34.10, z-score = -0.96, R) >consensus GCUUCCGCGUGGAUCUACCCAUCAAGAGCGCCCGUUACCGCGGCCAGUACAGCAGCUAUCCCAUCAAGCUGUUCUACACCUCCAACAUUCCCAUCAUUCUGCAGUCUGCGCUCGUCUCCA ........((((((...............(.((((....)))).).(((.(((((((.........))))))).)))..........................))))))........... (-18.92 = -18.21 + -0.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:23 2011