
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,318,871 – 2,318,985 |
| Length | 114 |
| Max. P | 0.536323 |

| Location | 2,318,871 – 2,318,985 |
|---|---|
| Length | 114 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.56605 |
| G+C content | 0.63045 |
| Mean single sequence MFE | -47.18 |
| Consensus MFE | -14.08 |
| Energy contribution | -13.82 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2318871 114 - 27905053 UCUUUUGGCAGCCGCC---GAGGCGAGAGUCUUGUACAGGUGGCUCUUGUUGGGCGGCAGACCGAUUACGCCGGAAGCGGAAGUGGUAGGAGGUGGCGAGGGAGGUGGUUCAGUAGG (((((((.((((((((---..((((((((((..........)))))))))).))))))...((.(((((.(((....)))..))))).))...)).))))))).............. ( -46.80, z-score = -2.64, R) >droSim1.chr3R 2347448 111 - 27517382 UCUUUUGGCAGCCGCC---GAGGCGAGUGUCUUAUACAGUUGACUCUUGUUGGGCGGCAGACCGAUUACGCCGGAAGCGGAAGUGCUUGGAGG---CGGGGGCGGUGGUUCAGUGGG .((.(((((.((((((---..((((((.(((..........))).)))))).))))))..((((....((((..(((((....)))))...))---))....)))).).)))).)). ( -44.20, z-score = -1.22, R) >droSec1.super_6 2405479 111 - 4358794 UCUUUUGGCAGCCGCC---GAGGCGAGUGUCUUGUACAGGUGACUCUUGUUGGGCGGCAGACCGAUUACGCCGGUAGCGGAAGUGGUUGGAGG---CGGGGGCGGUGGUUCAGUGGG .((.(((((.((((((---..((((((.(((..........))).)))))).))))))..((((....((((..((((.......))))..))---))....)))).).)))).)). ( -43.10, z-score = -0.83, R) >droYak2.chr3R 18238607 111 - 28832112 UCUUUUGGCAGCCGCC---GAGGCGAGUGUCUUGUACAGGUGACUCUUGUUGGGCGGCAGACCGAUUACGCCGGAAGCGGAAGUGGUUGGCGG---CGGGGGCGGUGGUUCCGUGGG ..........((((((---..((((((.(((..........))).)))))).))))))...(((....((((((..((....))..)))))).---)))..((((.....))))... ( -48.00, z-score = -1.35, R) >droEre2.scaffold_4770 2565017 111 - 17746568 UCUCUUGGCAGCCGCC---GAGGCGAGGGUCUUGUACAGGUGACUCUUGUUGGGCGGCAGACCGAUUACGCCAGAGGCGGAAGUGGUUGGAGG---CGGGGGCGGUGGUUCCGUGGG ...(((.((.((((((---..((((((((((..........)))))))))).)))))).(((((....((((...))))....)))))....)---).)))((((.....))))... ( -49.10, z-score = -1.70, R) >droAna3.scaffold_13340 13546802 99 - 23697760 GCGCUUGGCCGCCGCC---GAGGCCAGCGUCUUGUACAGCUGGCUCUUGUUGGGCGUAAG---------UCCGGCGAGGGAUGUUGGAGG------CGGAGGUGGUGGCUCUGAGGG .(.(((((((((((((---....((.((.(((...(((.....((((((((((((....)---------))))))))))).))).))).)------))).))))))))))..))).) ( -52.20, z-score = -2.86, R) >dp4.chr2 10731421 99 + 30794189 CCGCUUGGCCGCUGCC---GUGGCCAGGGUCUUGUACAGAUGACUCUUGUUCGGCGACA---------UGCCACUGACAGAUGUCGGUGG------CGGAGGCGGCGGCUCUGUGGG ((((..((((((((((---((.(((((((((..........)))))).....))).)).---------((((((((((....))))))))------))..))))))))))..)))). ( -60.20, z-score = -5.67, R) >droWil1.scaffold_181089 540750 114 - 12369635 UCUCUUUGCCGCCGCC---GAGGCCAGAGAUUUGUACAGGUGACUUUUAUUGGGCGAAAGACCCACUGAAGCGGUGGCUGCUCCUGAUGUAGGCGGUGGUGGCGGCGGUUCGGUUGG ...((..(((((((((---(..(((......((((..((....))((((.(((((....).)))).)))))))).(.((((.......)))).))))..))))))))))..)).... ( -46.40, z-score = -1.27, R) >droGri2.scaffold_14830 4599577 93 + 6267026 UCUUUUGGCUGCCGCCGCUGAGGCGAGUGAUUUGUACAGGUGGCUCUUGUUUGGAGAGAGUCC-----AGCUGGUGG-------------------UGCUGGUGGUGGCUCUGUAGG .((...(((..(((((((....)).(((.(((....(((.((((((((.......))))).))-----).)))..))-------------------)))))))))..)))....)). ( -34.60, z-score = -0.84, R) >consensus UCUUUUGGCAGCCGCC___GAGGCGAGUGUCUUGUACAGGUGACUCUUGUUGGGCGGCAGACCGAUUACGCCGGAGGCGGAAGUGGUUGGAGG___CGGGGGCGGUGGUUCAGUGGG ......(((.((((((...(((((....)))))....((....)).......................................................)))))).)))....... (-14.08 = -13.82 + -0.26)
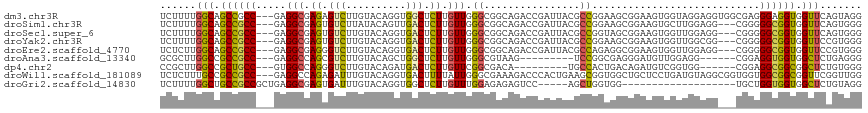
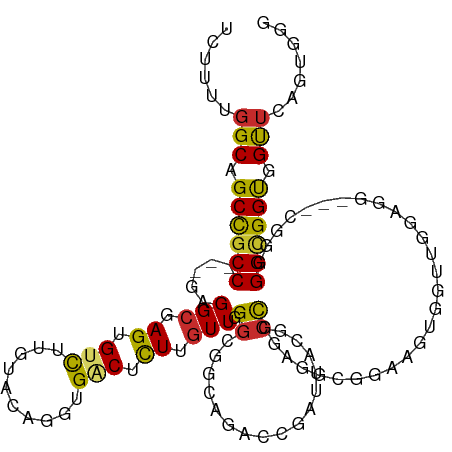
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:38 2011