| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,310,440 – 2,310,530 |
| Length | 90 |
| Max. P | 0.905957 |

| Location | 2,310,440 – 2,310,530 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.66440 |
| G+C content | 0.40000 |
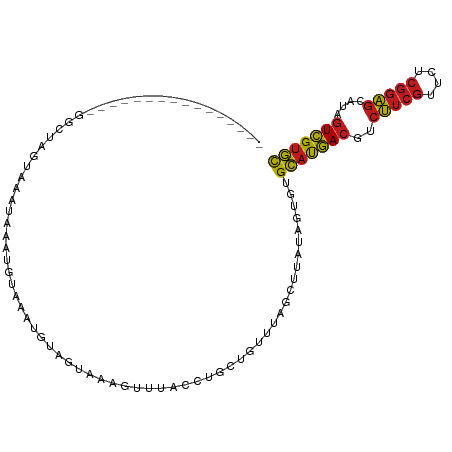
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.31 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
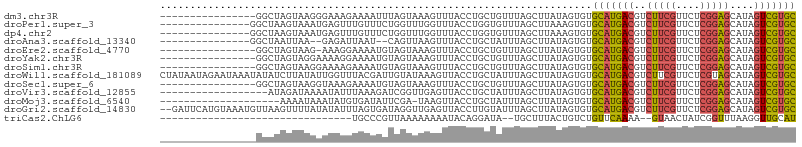
>dm3.chr3R 2310440 90 - 27905053 ----------------GGCUAGUAAGGGAAAGAAAAUUUAGUAAAGUUUACCUGCUGUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ----------------((((((..(((......((((((....)))))).))).....))))))........(((((((..(((((....)))))....))))))) ( -20.70, z-score = -0.25, R) >droPer1.super_3 4559398 91 + 7375914 ---------------GGCUAAGUAAAUGAGUUUGUUUCUGGUUUGGUUUACCUGGUGUUUAGCUUAAAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ---------------(((((((.((((.((.......)).))))((....)).....)))))))........(((((((..(((((....)))))....))))))) ( -22.40, z-score = -1.04, R) >dp4.chr2 10722645 91 + 30794189 ---------------GGCUAAGUAAAUGAGUUUGUUUCUGGUUUGGUUUACCUGGUGUUUAGCUUAAAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ---------------(((((((.((((.((.......)).))))((....)).....)))))))........(((((((..(((((....)))))....))))))) ( -22.40, z-score = -1.04, R) >droAna3.scaffold_13340 13538729 87 - 23697760 ---------------GGCUAAUUAA--GAGAUUAAU--CAGUUAAGUUUACCUGCUAUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ---------------.((((.....--((......)--)((((((((.........))))))))..))))..(((((((..(((((....)))))....))))))) ( -20.40, z-score = -0.69, R) >droEre2.scaffold_4770 2556133 89 - 17746568 ----------------GGCUAGUAAG-AAAGGAAAAUGUAGUAAAGUUUACCUGCUGUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ----------------((((((....-..(((.((((........)))).))).....))))))........(((((((..(((((....)))))....))))))) ( -23.30, z-score = -1.25, R) >droYak2.chr3R 18230081 90 - 28832112 ----------------GGCUAGUAGGAAAAGGAAAAUGUAGUAAAGUUUACCUGCUGUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ----------------((((((..((...(((.((((........)))).))).))..))))))........(((((((..(((((....)))))....))))))) ( -24.50, z-score = -1.50, R) >droSim1.chr3R 2338861 90 - 27517382 ----------------GGCUAGUAAGGAAAAGAAAAUGUAGUAAAGUUUACCUGCUGUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ----------------((((((...((...((.....((((......)))))).))..))))))........(((((((..(((((....)))))....))))))) ( -20.20, z-score = -0.22, R) >droWil1.scaffold_181089 530756 106 - 12369635 CUAUAAUAGAAUAAAUAUAUCUUAUAUUGGUUUACGAUUGUAUAAAGUUACCUGCUAUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGUAGCAUAGUCGUGC .((((((.((((.((((((...)))))).))))...))))))...........(((((.......)))))..(((((((..((.((....)).))....))))))) ( -16.90, z-score = 1.12, R) >droSec1.super_6 2396910 90 - 4358794 ----------------GGCUAGUAAGGUAAAGAAAAUGUAGUAAAGUUUACCUGCUGUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ----------------((((((..(((((((...............))))))).....))))))........(((((((..(((((....)))))....))))))) ( -23.96, z-score = -1.55, R) >droVir3.scaffold_12855 970493 88 + 10161210 ------------------AUAGAUAAAAUAUUUAAAGAUCGGUUGAGUUACCUGCUAUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ------------------...................((.((((((((........)))))))).)).....(((((((..(((((....)))))....))))))) ( -18.90, z-score = -0.24, R) >droMoj3.scaffold_6540 9852434 85 + 34148556 --------------------AAAAUAAAUAUGUGAUAUUCGA-UAAGUUACCUGCUAUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC --------------------..............((((...(-(((((((.........)))))))).))))(((((((..(((((....)))))....))))))) ( -19.20, z-score = -0.90, R) >droGri2.scaffold_14830 4590366 104 + 6267026 --GAUUCAUGUAAAUGUUAAGUUUUAUAUAUUUAGUGAUAGGUUGAGUUACCUUGUAUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC --...((((.(((((((..........)))))))))))((((((((((........))))))))))......(((((((..(((((....)))))....))))))) ( -25.10, z-score = -1.76, R) >triCas2.ChLG6 6159203 70 + 13544221 --------------------------------UGCCCGUUAAAAAAAAUACAGGAUA--UGCUUUACUGUCUGUUCAAAA--GUAACUAUCGGUUUAAGGUUGCAU --------------------------------....................(((((--.((......)).)))))....--((((((..........)))))).. ( -9.10, z-score = 0.48, R) >consensus ________________GGCUAGUAAAUAAAUGUAAAUGUAGUAAAGUUUACCUGCUGUUUAGCUUAUAGUGUGCAUGACGUCUUCGUUCUCGGAGCAUAGUCGUGC ........................................................................(((((((..(((((....)))))....))))))) (-12.41 = -12.31 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:37 2011