
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,265,854 – 2,265,946 |
| Length | 92 |
| Max. P | 0.847033 |

| Location | 2,265,854 – 2,265,946 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.35 |
| Shannon entropy | 0.74878 |
| G+C content | 0.60897 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.95 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847033 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 2265854 92 + 27905053 AGCGCAAAAAGUUGACUAGGAAGGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCCCAAAGGGAAGCGGAGGCGGCAGCGGCGGCGGCGGUA------------------- ..(((.............(((..(((((....).))))...)))(((((((((((((....(....)....).)))))))))))).)))...------------------- ( -38.90, z-score = -0.42, R) >droGri2.scaffold_14906 12484195 111 + 14172833 AGCGCAAGAAGCUCACACGUAAAGCGGCAAUUGCCAGCAAAUCGGCAGCAGCGGCGCAACGUGAAGCGGAAGAGGCAGCUGCUGUUGCAGCGGUUGCUGCGUCCGCCACAU (((.......)))..(((((...((.((..(((((........)))))..)).))...)))))..(((((...(((((((((((...)))))))))))...)))))..... ( -49.40, z-score = -1.54, R) >droMoj3.scaffold_6540 19907005 111 - 34148556 AGCGCAAGAAGCUAACACGCAAGGCGGCCAUUGCCAGCAAAUCGGCAGCGGCUGCGCAGCGGGAAGCUGAGGAGGCGGCCGCAGUCGCUGCUGUUGCUGCAUCCGCCACGU .(((......((......))..(((((....(((.(((((..(((((((((((((.((((.....)))).((......))))))))))))))))))))))).))))).))) ( -54.80, z-score = -1.20, R) >droVir3.scaffold_13047 17427114 111 + 19223366 AGCGGAAGAAGCUAACGCGCAAAGCGGCUAUUGCCAGCAAAUCGGCGGCGGCCGCACAGCGCGAAGCGGAAGAGGCAGCUGCUGUUGCUGCAGUUACUGCAUCCGCGACGU .(((((.........(((((...(((((..(((((........)))))..)))))...)))))..((((.((..(((((.......)))))..)).)))).)))))..... ( -54.10, z-score = -2.14, R) >droWil1.scaffold_181130 13098840 95 - 16660200 AGCGUAAAAAGCUAACACGCAAAGCAGCCAUUGCCAGUAAAUCAGCGGCUGCUGCCCAACGGGCUACUGCCGAGGCAGCGGCUGUUGCUGCUGCA---------------- .((((...........))))...(((((..(((....)))..((((((((((((((.....(((....)))..))))))))))))))..))))).---------------- ( -40.90, z-score = -1.17, R) >droSim1.chr3R 2292820 92 + 27517382 AGCGCAAAAAGUUGACAAGGAAGGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCCCAAAGGGAAGCGGAGGCGGCAGCGGCGGCGGCGUCA------------------- .((((.............(((..(((((....).))))...)))(((((((((((((....(....)....).)))))))))))).))))..------------------- ( -39.80, z-score = -0.85, R) >droSec1.super_6 2352646 92 + 4358794 AGCGCAAAAAGUUGACUAGGAAAGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCUCAAAGGGAAGCGGAGGCGGCAGCGGUGGCGGCGUCA------------------- .((((....((((.........))))......((((.(....((((((((.(((((........))))).)))))))).).)))).))))..------------------- ( -35.90, z-score = -0.29, R) >droEre2.scaffold_4770 2512181 92 + 17746568 AGCGCAAAAAGUUGACUAGGAAAGCUGCGAUAGCCAGCAAAUCUGCUGCCGCUGCACAAAGGGAAGCGGAGGCUGCAGCAGCGGCGGCGGCA------------------- ...((..................(((((....).)))).....((((((((((((.(....)...((((...)))).)))))))))))))).------------------- ( -35.00, z-score = 0.20, R) >dp4.chr2 10678678 90 - 30794189 AGCGGAAGAAACUCACCCGCAAAGCGGCAAUUGCCAGCAAGUCGGCGGCAGCGGCCCAGAGGGAGGCGGAGGCGGCCGCCGCCGUGGCUU--------------------- .((((..(.....)..)))).....(((....)))(((....(((((((.((.((((...(.....).).))).)).)))))))..))).--------------------- ( -39.60, z-score = 0.06, R) >droPer1.super_3 4515607 90 - 7375914 AGCGGAAGAAACUCACCCGCAAAGCGGCAAUUGCCAGCAAGUCGGCGGCAGCGGCCCAGAGGGAGGCGGAGGCGGCCGCCGCCGUGGCUU--------------------- .((((..(.....)..)))).....(((....)))(((....(((((((.((.((((...(.....).).))).)).)))))))..))).--------------------- ( -39.60, z-score = 0.06, R) >triCas2.ChLG5 10965479 71 - 18847211 AGUGGGGAACAAUAAUUA--AUUCUUGCGGUU-UUAAAGAAUGGAAAGCGACAGCCGAAGAAAUUACGGAGAAA------------------------------------- .(..((((..........--.))))..)(.((-((........)))).).....(((((....)).))).....------------------------------------- ( -9.50, z-score = 0.08, R) >consensus AGCGCAAAAAGCUAACACGCAAAGCGGCGAUUGCCAGCAAAUCGGCGGCCGCUGCCCAAAGGGAAGCGGAGGCGGCAGCGGCGGUGGCUGCA___________________ .((..............((.....))((....))..))....(((((((((((((...................)))))))))))))........................ (-19.30 = -19.35 + 0.05)
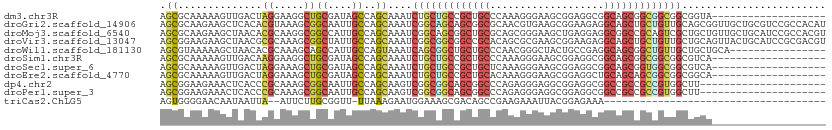

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:34 2011