
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,185,082 – 2,185,177 |
| Length | 95 |
| Max. P | 0.665255 |

| Location | 2,185,082 – 2,185,177 |
|---|---|
| Length | 95 |
| Sequences | 14 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Shannon entropy | 0.55037 |
| G+C content | 0.50189 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -13.63 |
| Energy contribution | -12.96 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
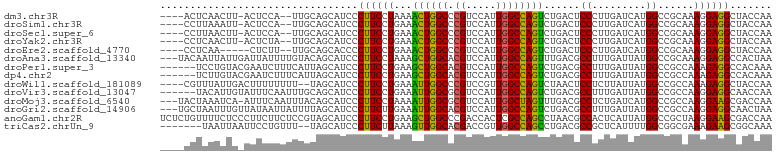
>dm3.chr3R 2185082 95 + 27905053 ----ACUCAACUU-ACUCCA--UUGCAGCAUCCCUUCCUAAAACUGGCCCGUCCAUUGGCCAGUCUGACUCCCUUGAUCAUGGCCGCAAAGGAGGCUACCAA ----.........-.((((.--((((.((.............(((((((........))))))).((((......).)))..)).)))).))))........ ( -26.20, z-score = -2.09, R) >droSim1.chr3R 2212164 95 + 27517382 ----CCUUAAAUU-ACUCCA--UUGCAGCAUCCCUUCCUGAAACUGGCCCGUCCAUUGGCCAGUCUGACUCCCUUGAUCAUGGCCGCAAAGGAGGCUACCAA ----.........-.((((.--((((.((.............(((((((........))))))).((((......).)))..)).)))).))))........ ( -26.20, z-score = -1.63, R) >droSec1.super_6 2270246 95 + 4358794 ----CCUUAACUU-ACUCCA--UUGCAGCAUCCCUUCCUGAAACUGGCCCGUCCAUUGGCCAGUCUGACUCCCUUGAUCAUGGCCGCAAAGGAGGCUACCAA ----.........-.((((.--((((.((.............(((((((........))))))).((((......).)))..)).)))).))))........ ( -26.20, z-score = -1.63, R) >droYak2.chr3R 18110531 95 + 28832112 ----CCUCAACUU-ACUCUA--UUGCAGCAUCCCUUCCUGAAACUGGCCCGUCCAUUGGCCAGUCUGACUCCCUUGAUCAUGGCCGCAAAGGAGGCUACCAA ----.........-.((((.--((((.((.............(((((((........))))))).((((......).)))..)).)))).))))........ ( -24.40, z-score = -1.00, R) >droEre2.scaffold_4770 2429594 91 + 17746568 ----CCUCAA-----CUCUU--UUGCAGCACCCCUUCCUGAAACUGGCCCGUCCAUUGGCCAGUCUGACUCCCUUGAUCAUGGCCGCAAAGGAGGCUACCAA ----......-----(((((--((((.((.............(((((((........))))))).((((......).)))..)).)))))))))........ ( -27.60, z-score = -1.95, R) >droAna3.scaffold_13340 13420659 99 + 23697760 ---UACAAUUAUUGAUUAUUUUGUACAGCAUCCCUUCCUAAAGCUGGCACGUCCAUUGGCCAGUUUGACGCCUUUGAUUAUGGCCGCAAAGGAGGCCACUAA ---(((((............)))))........((((((.((((((((.((.....))))))))))...(((.........))).....))))))....... ( -23.20, z-score = -0.33, R) >droPer1.super_3 4422214 96 - 7375914 ------UCCUGUACGAAUCUUUCAUUAGCAUCCCUUCCUGAAGCUGGCACGUCCAUUGGCCAGUCUGACGCCUUUGAUUAUGGCCGCCAAAGAGGCCACAAA ------.........((((.((((..((.....))...))))((((((.((.....))))))))...........)))).(((((........))))).... ( -20.40, z-score = 0.61, R) >dp4.chr2 10585442 96 - 30794189 ------UCUUGUACGAAUCUUUCAUUAGCAUCCCUUCCUGAAGCUGGCACGUCCAUUGGCCAGUCUGACGCCUUUGAUUAUGGCCGCCAAAGAGGCCACAAA ------..((((...((((.((((..((.....))...))))((((((.((.....))))))))...........))))..((((........)))))))). ( -21.20, z-score = 0.34, R) >droWil1.scaffold_181089 10610863 96 + 12369635 ----CGUUUAUUGACUUUUUUUU--UAGCAUCCCUUCCUGAAAUUGGCCCGUCCGUUGGCCAGUCUAACUCCUCUUAUUAUGGCCGCCAAAGAGGCUACCAA ----.(((....(((.....(((--(((.(.....).)))))).(((((........)))))))).)))...........(((..(((.....)))..))). ( -19.50, z-score = -0.58, R) >droVir3.scaffold_13047 17342461 96 + 19223366 ------UACAUUGUAUUUCAAUUUGCAGCAUCCCUUCCUGAAAUUGGCGCGUCCAUUGGCCAGUCUGACGCCUUUGAUUAUGGCCGCCAAGGAGGCAACCAA ------...........(((((((.(((.(.....).)))))))))).((.(((.(((((..(.....)(((.........))).)))))))).))...... ( -23.00, z-score = 0.41, R) >droMoj3.scaffold_6540 19832095 98 - 34148556 ---UACUAAAUCA-AUUUCAAUUUACAGCAUCCCUUCCUAAAAUUGGCGCGUCCAUUGGCUAGUUUGACGCCUCUGAUCAUGGCCGCCAAGGAAGCGACCAA ---..........-.............................(((((((.(((.(((((..(.....)(((.........))).)))))))).))).)))) ( -22.60, z-score = -1.05, R) >droGri2.scaffold_14906 12400057 99 + 14172833 ---UGCUAAUUUGUUAUAAUUAUUUUAGCAUCCCUUCUUGAAAUUGGCACGUCCAUUGGCCAGUCUGACGCCUUUGAUUAUGGCCGCCAAGGAGGCAACUAA ---((((((..((.......))..))))))..((((((((..((((((.((.....)))))))).....(((.........)))...))))))))....... ( -26.80, z-score = -1.53, R) >anoGam1.chr2R 32158018 102 - 62725911 UCUCUGUUUUCUCCCUUCUUCUCCGUAGCAUCCCUUCCUGAAGCUGGCCCGACCACUCGCCAGCCUAACGCCACUCAUUAUGGCCGCUAAGGAAGCGACCAA ...........((.((((((.....((((.............((((((..........)))))).....((((.......)))).)))))))))).)).... ( -23.10, z-score = -1.13, R) >triCas2.chrUn_9 742288 93 - 915065 -------UAAUUAAUUCCUGUUU--UAGCAUCCCUUCUUAAAGUUGGCACGACCGUUGGCCAGCCUGACGCCGCUCAUUUUGGCGGCGAAAGAAGCGGCAAA -------................--..((....((((((...((((((.(((...)))))))))....(((((((......))))))).))))))..))... ( -29.80, z-score = -1.57, R) >consensus ____CCUCAAUUU_AUUCCUUUUUGCAGCAUCCCUUCCUGAAACUGGCCCGUCCAUUGGCCAGUCUGACGCCUUUGAUUAUGGCCGCAAAGGAGGCUACCAA .................................((((((...((((((.((.....))))))))......((.........))......))))))....... (-13.63 = -12.96 + -0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:28 2011