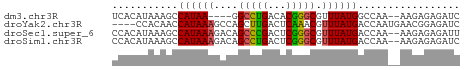
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,150,123 – 2,150,175 |
| Length | 52 |
| Max. P | 0.971849 |

| Location | 2,150,123 – 2,150,175 |
|---|---|
| Length | 52 |
| Sequences | 4 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.37344 |
| G+C content | 0.46345 |
| Mean single sequence MFE | -12.47 |
| Consensus MFE | -9.99 |
| Energy contribution | -9.68 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
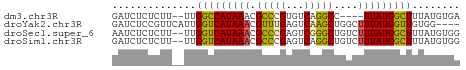
>dm3.chr3R 2150123 52 + 27905053 GAUCUCUCUU--UUGGCCAUAAACGCCCGUGUCAGGCC----UUAUGGCUUUAUGUGA ....((.(..--..((((((((..(((.......))).----))))))))....).)) ( -13.70, z-score = -1.72, R) >droYak2.chr3R 18077594 54 + 28832112 GAUCUCCGUUCAUUGGUCAUAAACGUUUGAGUCAAGCUGGCUUUAUGGUUGUGG---- .....(((.....))).(((((.(((..(((((.....))))).))).))))).---- ( -10.30, z-score = -0.33, R) >droSec1.super_6 2235759 56 + 4358794 AAUCUCUCUU--UUGGUCAUAAACGCCCGAGUCGGGCUGUCUUUAUGGCUUUAUGUGG .....(.(..--..(((((((((((((((...))))).))..))))))))....).). ( -15.30, z-score = -2.44, R) >droSim1.chr3R 2176014 56 + 27517382 GAUCUCUCUU--UUGGUCAUAAACGCCCGAGUCAGGCUGUCUUUAUGGCUUUAUGUGG .....(.(..--..(((((((((.((((......))..)).)))))))))....).). ( -10.60, z-score = -0.21, R) >consensus GAUCUCUCUU__UUGGUCAUAAACGCCCGAGUCAGGCUGUCUUUAUGGCUUUAUGUGG ..............(((((((((.(((((...)))))....)))))))))........ ( -9.99 = -9.68 + -0.31)
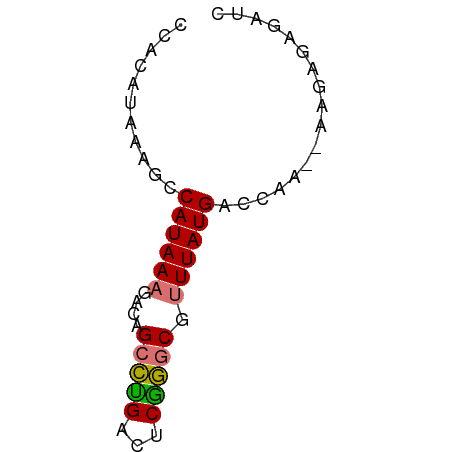
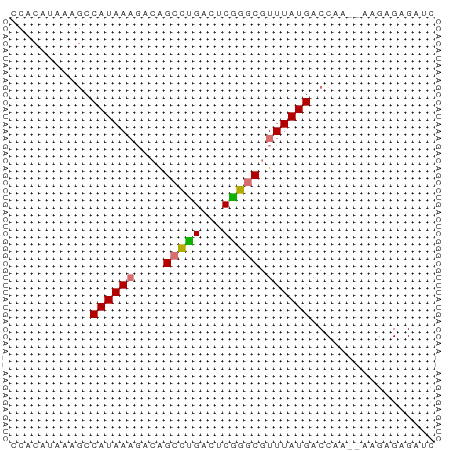
| Location | 2,150,123 – 2,150,175 |
|---|---|
| Length | 52 |
| Sequences | 4 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.37344 |
| G+C content | 0.46345 |
| Mean single sequence MFE | -11.30 |
| Consensus MFE | -7.80 |
| Energy contribution | -7.55 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
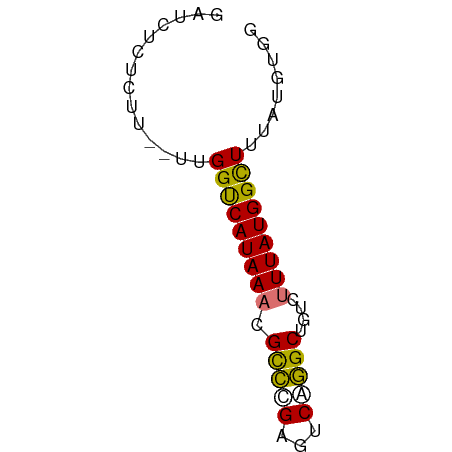
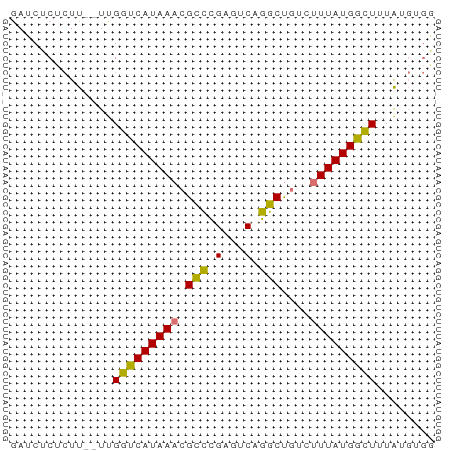
>dm3.chr3R 2150123 52 - 27905053 UCACAUAAAGCCAUAA----GGCCUGACACGGGCGUUUAUGGCCAA--AAGAGAGAUC .........(((((((----((((((...))))).))))))))...--.......... ( -16.50, z-score = -3.53, R) >droYak2.chr3R 18077594 54 - 28832112 ----CCACAACCAUAAAGCCAGCUUGACUCAAACGUUUAUGACCAAUGAACGGAGAUC ----...........(((....)))..(((...(((((((.....))))))))))... ( -7.90, z-score = -1.33, R) >droSec1.super_6 2235759 56 - 4358794 CCACAUAAAGCCAUAAAGACAGCCCGACUCGGGCGUUUAUGACCAA--AAGAGAGAUU ...........((((((....(((((...))))).)))))).....--.......... ( -11.50, z-score = -2.77, R) >droSim1.chr3R 2176014 56 - 27517382 CCACAUAAAGCCAUAAAGACAGCCUGACUCGGGCGUUUAUGACCAA--AAGAGAGAUC ...........((((((....(((((...))))).)))))).....--.......... ( -9.30, z-score = -1.13, R) >consensus CCACAUAAAGCCAUAAAGACAGCCUGACUCGGGCGUUUAUGACCAA__AAGAGAGAUC ...........((((((....(((((...))))).))))))................. ( -7.80 = -7.55 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:26 2011