| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,138,497 – 2,138,608 |
| Length | 111 |
| Max. P | 0.983003 |

| Location | 2,138,497 – 2,138,608 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.89 |
| Shannon entropy | 0.49457 |
| G+C content | 0.39418 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -13.31 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
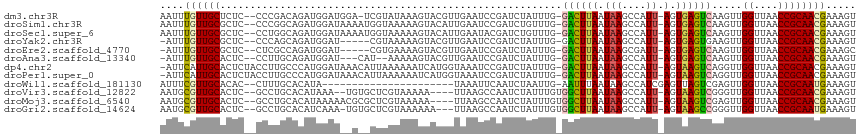
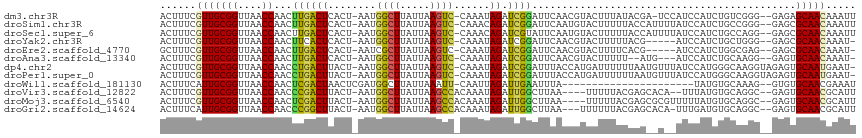
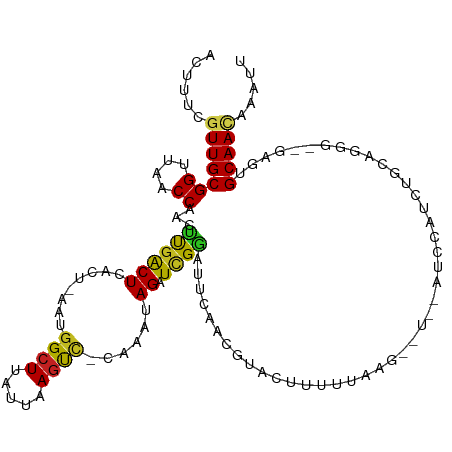
>dm3.chr3R 2138497 111 + 27905053 AAUUUGUUGCUCUC--CCCGACAGAUGGAUGGA-UCGUAUAAAGUACGUUGAAUCCGAUCUAUUUG-GACUUAAUAAGCCAUU-AGUGAGUCAAGUUGGUUAACCGCAACGAAAGU ..((((((((....--.((((((((((((((((-((((((...)))))....)))).)))))))).-((((((.(((....))-).))))))..)))))......))))))))... ( -34.10, z-score = -3.58, R) >droSim1.chr3R 2132362 112 + 27517382 AAUUUGUUGCGCUC--CCCGGCAGAUGGAUAAAAUGGUAAAAAGUACAUUGAAUCCGAUCUGUUUG-GACUUAAUAAGCCAUU-AGUGAGUCAAGUUGGUUAACCGCAACGAAAGU ..(((((((((...--.(((((((((((((..((((..........))))..)))).)))).....-((((((.(((....))-).))))))..))))).....)))))))))... ( -33.50, z-score = -3.29, R) >droSec1.super_6 2224275 112 + 4358794 AAUUUGUUGCGCUC--CCUGGCAGAUGGAUAAAAUGGUAAAAAGUACAUUGAAUACGAUCUGUUUG-GACUUAAUAAGCCAUU-AGUGAGUCAAGUUGGUUAACCGCAACGAAAGU ..(((((((((..(--((..((((((.(((..((((..........))))..)).).))))))..)-((((((.(((....))-).)))))).....)).....)))))))))... ( -29.00, z-score = -2.02, R) >droYak2.chr3R 18066175 106 + 28832112 -AUUUGUUGCGCUC--CCCAGCAGAUGGAU-----CGUAAAAAGUACGUUGAAUCCGAUCUAUUUG-GACUUAAUAAGCCAUU-AGUGAGUGAAGUUGGUUAACCGCAACGAAAGU -.(((((((((...--.(((((((((((((-----((..................)))))))))).-.(((((.(((....))-).)))))...))))).....)))))))))... ( -30.07, z-score = -2.90, R) >droEre2.scaffold_4770 2383768 106 + 17746568 -AUUUGUUGCGCUC--CUCGCCAGAUGGAU-----CGUGAAAAGUACGUUGAAUCCGAUCUAUUUG-GACUUAAUAAGCGAUU-AGUGAGUCAAGUUGGUUAACCGCAACGAAAGC -.(((((((((..(--(....(((((((((-----((.((.............)))))))))))))-((((((.(((....))-).)))))).....)).....)))))))))... ( -32.32, z-score = -2.72, R) >droAna3.scaffold_13340 5673397 106 - 23697760 -AUUUGUUGCACUC--CCUUGCAGAUGGAU---CAU--AAAAAGUACGUUGAAUCCGAUCUAUUUG-GACUUAAUAAGCCAUU-AGUGAGUCAAGUUGGUUAACCGCAACGAAAGU -.((((((((....--.(((.(((((((((---(..--..................))))))))))-((((((.(((....))-).)))))))))..((....))))))))))... ( -27.85, z-score = -2.51, R) >dp4.chr2 15098939 113 - 30794189 -AUUCAUUGCACUCUACCUUGCCCAUGGAUAAACAUUAAAAAAUCAUGGUAAAUCCGAUCUAUUUG-GACUUAAUAAGCCAUU-AGUAAGUCAGGUUGGUUAACCGCAACGAAAGU -.(((.((((......((........))...................(((.((.(((((((.....-((((((.(((....))-).))))))))))))))).))))))).)))... ( -24.10, z-score = -1.41, R) >droPer1.super_0 4719116 113 + 11822988 -AUUCAUUGCACUCUACCUUGCCCAUGGAUAAACAUUAAAAAAUCAUGGUAAAUCCGAUCUAUUUG-GACUUAAUAAGCCAUU-AGUAAGUCAGGUUGGUUAACCGCAACGAAAGU -.(((.((((......((........))...................(((.((.(((((((.....-((((((.(((....))-).))))))))))))))).))))))).)))... ( -24.10, z-score = -1.41, R) >droWil1.scaffold_181130 5493267 91 - 16660200 AUUUCGUUGCACAC--CUUUGCACAUA----------------------UAAAUUCAAUCUAAUUG-AAUUUAAUAAGCCAUCGAGUUAGUCGAGUUGGUUAACCGCAAUGAAAGU .(((((((((....--...........----------------------((((((((((...))))-))))))...(((((((((.....))))..)))))....))))))))).. ( -24.40, z-score = -4.15, R) >droVir3.scaffold_12822 2375038 107 - 4096053 AAUGCGUUGCACUC--GCCUGCACAUAAA--UGUGCUCGUAAAAA----UUAAGCCAAUCUAUUUGUGGCUUAAUAAGCCAUU-AGUAAGUCGGGUUGGUUAACCGCAACGAAAGU ....((((((....--((..(((((....--)))))..)).....----...(((((((((((((((((((.....)))))).-...)))).)))))))))....))))))..... ( -34.00, z-score = -3.25, R) >droMoj3.scaffold_6540 11418006 109 + 34148556 AAUGCGUUGCACUC--GCCUGCACAUAAAAACGCGCUCGUAAAAA----UUAAGCCAAUCUAUUUGUGGCUUAAUAAGCCAUU-AGUAAGUCGAGUUGGUUAACCGCAACGAAAGU ....((((((((((--((.(((........(((....)))....(----((((((((.........)))))))))........-.))).).))))).((....))))))))..... ( -28.80, z-score = -1.75, R) >droGri2.scaffold_14624 1494814 109 - 4233967 AAUGCGUUGCACUC--GCCUGCACAUCAAA-UGUGCUCGUAAAAAA---UUAAGCCAAUCUAUUUGUGGCUUAAUAAGCCAUU-AGUAAGCCGGGUUGGUUAACCGCAAUGAAAGU ....((((((....--((..((((((...)-)))))..))......---...((((((((((((.((((((.....)))))).-))).....)))))))))....))))))..... ( -32.20, z-score = -2.40, R) >consensus AAUUCGUUGCACUC__CCCUGCACAUGGAU__A__CGUAAAAAGUACGUUGAAUCCGAUCUAUUUG_GACUUAAUAAGCCAUU_AGUGAGUCAAGUUGGUUAACCGCAACGAAAGU ....((((((.........................................................((((((..((....))...)))))).....((....))))))))..... (-13.31 = -12.90 + -0.41)
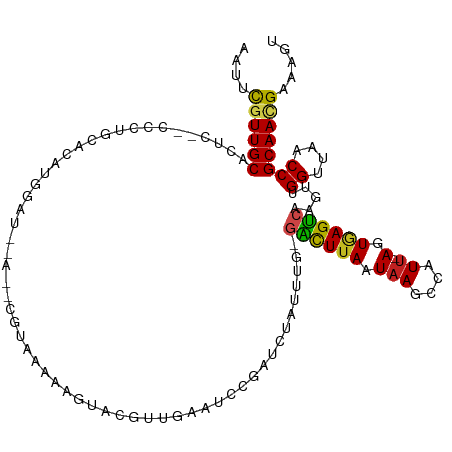
| Location | 2,138,497 – 2,138,608 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.89 |
| Shannon entropy | 0.49457 |
| G+C content | 0.39418 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -10.94 |
| Energy contribution | -10.31 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2138497 111 - 27905053 ACUUUCGUUGCGGUUAACCAACUUGACUCACU-AAUGGCUUAUUAAGUC-CAAAUAGAUCGGAUUCAACGUACUUUAUACGA-UCCAUCCAUCUGUCGGG--GAGAGCAACAAAUU ......(((((((....))..((((((.....-..(((((.....)).)-))...((((.((((....((((.....)))).-...))))))))))))))--....)))))..... ( -27.40, z-score = -2.23, R) >droSim1.chr3R 2132362 112 - 27517382 ACUUUCGUUGCGGUUAACCAACUUGACUCACU-AAUGGCUUAUUAAGUC-CAAACAGAUCGGAUUCAAUGUACUUUUUACCAUUUUAUCCAUCUGCCGGG--GAGCGCAACAAAUU ......((((((.((..((.............-..(((((.....)).)-))..(((((.((((..((((..........))))..)))))))))..)).--.))))))))..... ( -26.70, z-score = -2.33, R) >droSec1.super_6 2224275 112 - 4358794 ACUUUCGUUGCGGUUAACCAACUUGACUCACU-AAUGGCUUAUUAAGUC-CAAACAGAUCGUAUUCAAUGUACUUUUUACCAUUUUAUCCAUCUGCCAGG--GAGCGCAACAAAUU ......(((((((....)).....(.(((.((-..((((.......(((-......))).((((.....)))).....................))))))--)))))))))..... ( -19.50, z-score = -0.67, R) >droYak2.chr3R 18066175 106 - 28832112 ACUUUCGUUGCGGUUAACCAACUUCACUCACU-AAUGGCUUAUUAAGUC-CAAAUAGAUCGGAUUCAACGUACUUUUUACG-----AUCCAUCUGCUGGG--GAGCGCAACAAAU- ......((((((.((..(((............-..(((((.....)).)-))..(((((.((((....((((.....))))-----))))))))).))).--.))))))))....- ( -25.20, z-score = -2.26, R) >droEre2.scaffold_4770 2383768 106 - 17746568 GCUUUCGUUGCGGUUAACCAACUUGACUCACU-AAUCGCUUAUUAAGUC-CAAAUAGAUCGGAUUCAACGUACUUUUCACG-----AUCCAUCUGGCGAG--GAGCGCAACAAAU- ......(((((((((((.....))))))....-....((((.....(.(-((.((.(((((((.............)).))-----))).)).))))...--)))))))))....- ( -24.82, z-score = -1.69, R) >droAna3.scaffold_13340 5673397 106 + 23697760 ACUUUCGUUGCGGUUAACCAACUUGACUCACU-AAUGGCUUAUUAAGUC-CAAAUAGAUCGGAUUCAACGUACUUUUU--AUG---AUCCAUCUGCAAGG--GAGUGCAACAAAU- ......(((((((....))......((((.((-..(((((.....)).)-))..(((((.((((....((((.....)--)))---)))))))))..)).--)))))))))....- ( -23.20, z-score = -1.38, R) >dp4.chr2 15098939 113 + 30794189 ACUUUCGUUGCGGUUAACCAACCUGACUUACU-AAUGGCUUAUUAAGUC-CAAAUAGAUCGGAUUUACCAUGAUUUUUUAAUGUUUAUCCAUGGGCAAGGUAGAGUGCAAUGAAU- ...(((((((((.((.(((..(((((((((.(-((....))).))))))-..........((((..((..(((....)))..))..))))..)))...))).)).))))))))).- ( -28.50, z-score = -1.95, R) >droPer1.super_0 4719116 113 - 11822988 ACUUUCGUUGCGGUUAACCAACCUGACUUACU-AAUGGCUUAUUAAGUC-CAAAUAGAUCGGAUUUACCAUGAUUUUUUAAUGUUUAUCCAUGGGCAAGGUAGAGUGCAAUGAAU- ...(((((((((.((.(((..(((((((((.(-((....))).))))))-..........((((..((..(((....)))..))..))))..)))...))).)).))))))))).- ( -28.50, z-score = -1.95, R) >droWil1.scaffold_181130 5493267 91 + 16660200 ACUUUCAUUGCGGUUAACCAACUCGACUAACUCGAUGGCUUAUUAAAUU-CAAUUAGAUUGAAUUUA----------------------UAUGUGCAAAG--GUGUGCAACGAAAU ..((((.(((((....(((...((((.....))))..((.(((((((((-((((...))))))))))----------------------.))).))...)--)).))))).)))). ( -20.90, z-score = -2.45, R) >droVir3.scaffold_12822 2375038 107 + 4096053 ACUUUCGUUGCGGUUAACCAACCCGACUUACU-AAUGGCUUAUUAAGCCACAAAUAGAUUGGCUUAA----UUUUUACGAGCACA--UUUAUGUGCAGGC--GAGUGCAACGCAUU .....((((((((((....))))..(((..((-..((....(((((((((.........))))))))----).....)).(((((--....))))).)).--.))))))))).... ( -30.10, z-score = -2.69, R) >droMoj3.scaffold_6540 11418006 109 - 34148556 ACUUUCGUUGCGGUUAACCAACUCGACUUACU-AAUGGCUUAUUAAGCCACAAAUAGAUUGGCUUAA----UUUUUACGAGCGCGUUUUUAUGUGCAGGC--GAGUGCAACGCAUU .....((((((((....)).(((((.(((...-..((....(((((((((.........))))))))----).....)).((((((....))))))))))--)))))))))).... ( -34.30, z-score = -3.20, R) >droGri2.scaffold_14624 1494814 109 + 4233967 ACUUUCAUUGCGGUUAACCAACCCGGCUUACU-AAUGGCUUAUUAAGCCACAAAUAGAUUGGCUUAA---UUUUUUACGAGCACA-UUUGAUGUGCAGGC--GAGUGCAACGCAUU .......((((((....)).((.((.(((((.-(((((((((((((((((.........))))))))---).......)))).))-))....))).)).)--).))))))...... ( -26.21, z-score = -0.90, R) >consensus ACUUUCGUUGCGGUUAACCAACUUGACUCACU_AAUGGCUUAUUAAGUC_CAAAUAGAUCGGAUUCAACGUACUUUUUAAG__U__AUCCAUCUGCAGGG__GAGUGCAACAAAUU ......(((((((....))...((((((........((((.....))))......)).))))............................................)))))..... (-10.94 = -10.31 + -0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:25 2011