| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,122,918 – 2,123,022 |
| Length | 104 |
| Max. P | 0.795685 |

| Location | 2,122,918 – 2,123,022 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.80 |
| Shannon entropy | 0.43302 |
| G+C content | 0.54372 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.66 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
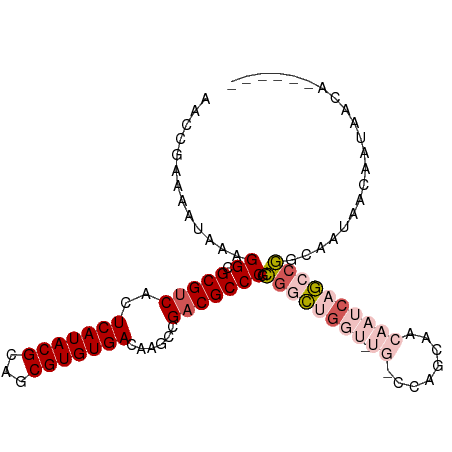
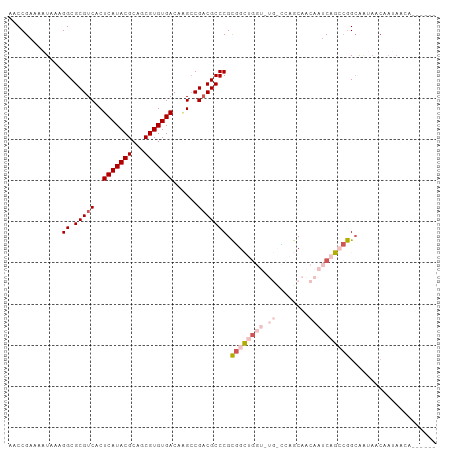
>dm3.chr3R 2122918 104 + 27905053 AACCGAAAAUAAAGGCGCGUCACUCAUACGCAUCGUGUGACAAGCCGACGCCCGCGGCUGGUAUGCCCAGCAACAAUCAGCCGGCAAUAACAAUAACAAUAAGC .............((.(((((..(((((((...)))))))......)))))))(((((((((.(((...)))...))))))).))................... ( -29.40, z-score = -1.49, R) >droSec1.super_6 2209049 104 + 4358794 AACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGCCGCCCGCGGCUGGUUUGCCCAGCAACAAUCAGCCGGCAAUAACAAUAACAAUAAGC .............((((.((...(((((((...)))))))...))))))(((...(((((((((((...)))..)))))))))))................... ( -31.60, z-score = -1.32, R) >droEre2.scaffold_4770 2367390 98 + 17746568 AACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCCCGCGGCUGGUUUGUCGAGCAACAAUCAGCCGGCAAUAACAAUAAGC------ .............((.(((((..(((((((...)))))))......)))))))((((((((((.((......)))))))))).)).............------ ( -29.90, z-score = -0.92, R) >droMoj3.scaffold_6540 11395401 92 + 34148556 AACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAUGGCGACGCCCGUAGGCG------UCAGCCAAAUUCACAGAGCAGAAACCAAAAGC------ .............((.((((.......)))).((.(((((..((((((((((....))))------)).))))...)))))..)).....))......------ ( -30.90, z-score = -3.25, R) >droGri2.scaffold_14624 1481401 80 - 4233967 AACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAGGCCGACGCCCGUGGGC-------UCCGCUAAAUCCACACAGAGC----------------- ..............((((((.......)))).))(((((...(((.((.(((....)))-------)).))).....))))).....----------------- ( -22.20, z-score = -0.11, R) >consensus AACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCCCGCGGCUGGU_UG_CCAGCAACAAUCAGCCGGCAAUAACAAUAACA______ .............((.(((((..(((((((...)))))))......))))))).((((((((.............))))))))..................... (-18.42 = -19.66 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:20 2011