| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,107,705 – 2,107,795 |
| Length | 90 |
| Max. P | 0.719232 |

| Location | 2,107,705 – 2,107,795 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Shannon entropy | 0.47043 |
| G+C content | 0.50101 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -15.40 |
| Energy contribution | -14.64 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2107705 90 + 27905053 CAACGGUUCACUCGACGCACUGUG-GCACCUAUGGGUACAUCGGAUGAAAUGUAUCCAGA--ACGUGCGUUCCCACUGGG----CACUGCGGAUUUU------------- ...(((.....))).((((..(((-((((...(((((((((........)))))))))..--..))))...(((...)))----)))))))......------------- ( -25.90, z-score = -0.19, R) >droSim1.chr3R 2121619 90 + 27517382 CAACGGUUCACUCGACGCACUGUG-GCACCUAUGGGUACAUCGGAUGAAAUGUAUCUAGA--ACGUGCGUUCCCACUGGG----CACUGCGGAUUUU------------- ...(((.....))).((((..(((-((((...(((((((((........)))))))))..--..))))...(((...)))----)))))))......------------- ( -23.60, z-score = 0.21, R) >droSec1.super_6 2193512 90 + 4358794 CAACGGUUCACUCGACGCACUGUG-GCACCUAUGGGUACAUCGGAUGAAAUGUAUCCAGA--ACGUGCGUUCCCACUGGG----CACUGCGGAUUUU------------- ...(((.....))).((((..(((-((((...(((((((((........)))))))))..--..))))...(((...)))----)))))))......------------- ( -25.90, z-score = -0.19, R) >droYak2.chr3R 18033701 88 + 28832112 CAACGGUUUAC-CGAUGCACUGUG-GCACCUAUGGGUACAUCGUAUAAAAUGUACCCAGA--ACGUGCGUUCCCACUGGG----CACUGCGGACUU-------------- ...(((....)-)).....((((.-((((...(((((((((........)))))))))..--..))))((.(((...)))----.)).))))....-------------- ( -29.80, z-score = -1.67, R) >droEre2.scaffold_4770 2350954 89 + 17746568 CAACGGUUCACUCGACGCACUGUG-GCACCUAUGGGUACAUCGGAUAAAAUGUAUCCAGA--ACGUGCGUUCCCACUGGG----CACUGCGGAUUU-------------- ...(((.....))).((((..(((-((((...(((((((((........)))))))))..--..))))...(((...)))----))))))).....-------------- ( -25.90, z-score = -0.52, R) >droAna3.scaffold_13340 5644300 90 - 23697760 CAACGGUUAACUCUACGCGCAGUG-GCACUUAUGGAUACAUCCACUGAAAUGUAUCUAGA--ACGUGCGUUCCCACUGGG----CAAUGCGGAUGAU------------- ................((.(((((-(......(((((((((........)))))))))((--((....)))))))))).)----)............------------- ( -26.40, z-score = -1.50, R) >dp4.chr2 15072082 100 - 30794189 CAACGGUUAACUCUCCGCCCAGUG-GCACCUAUGGAUACAUCGCCUGAAAUGUAUCUAGA--ACGUGCGUUCCCACUGUGGCCGCAAUGCAGUUCUUUGCACA------- ....((........))((((((((-(......(((((((((........)))))))))((--((....)))))))))).))).....(((((....)))))..------- ( -31.80, z-score = -2.24, R) >droPer1.super_0 4691862 100 + 11822988 CAACGGUUAACUCUCCUCCCAGUG-GCACCUAUGGAUACAUCGCCUGAAAUGUAUCUAGA--ACGUGCGUUCCCACUGUGGCCGCAAUGCAGUUCUUUGCACA------- ...((((((..........(((((-(......(((((((((........)))))))))((--((....))))))))))))))))...(((((....)))))..------- ( -28.50, z-score = -1.67, R) >droWil1.scaffold_181130 5457521 105 - 16660200 CAACGGUUUUCUCUACGCACAGUUUGCACCUUUGGAUACAUCGUCUGGAAUGUAUCUAUAUAGCGUGCAUUCACACUGUG-----ACCGCAUAUCAUAUCAGCAACUAUU ...((((..........((((((.(((((((.(((((((((........)))))))))...)).))))).....))))))-----))))..................... ( -26.00, z-score = -2.14, R) >droGri2.scaffold_14624 1464171 74 - 4233967 CAACGGUUA---------ACAGUU-GCACCUACGGAUACAUCGCUGGAAAUGUAUCUGUA--ACGUGCACUU--AGUG--------CCUUCUAUUU-------------- ....(((..---------..(((.-((((.(((((((((((........)))))))))))--..))))))).--...)--------))........-------------- ( -23.40, z-score = -3.34, R) >consensus CAACGGUUAACUCGACGCACAGUG_GCACCUAUGGAUACAUCGGAUGAAAUGUAUCUAGA__ACGUGCGUUCCCACUGGG____CACUGCGGAUUUU_____________ .........................((((...(((((((((........)))))))))......)))).......................................... (-15.40 = -14.64 + -0.76)

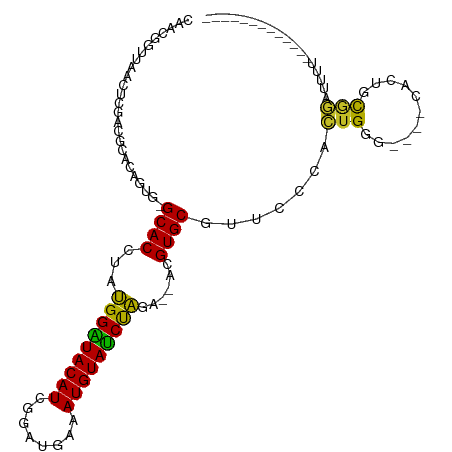
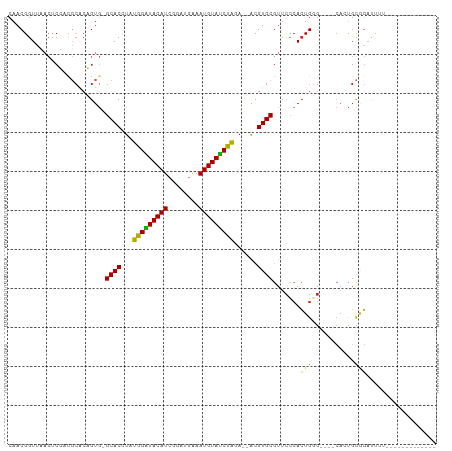
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:18 2011